Abstract
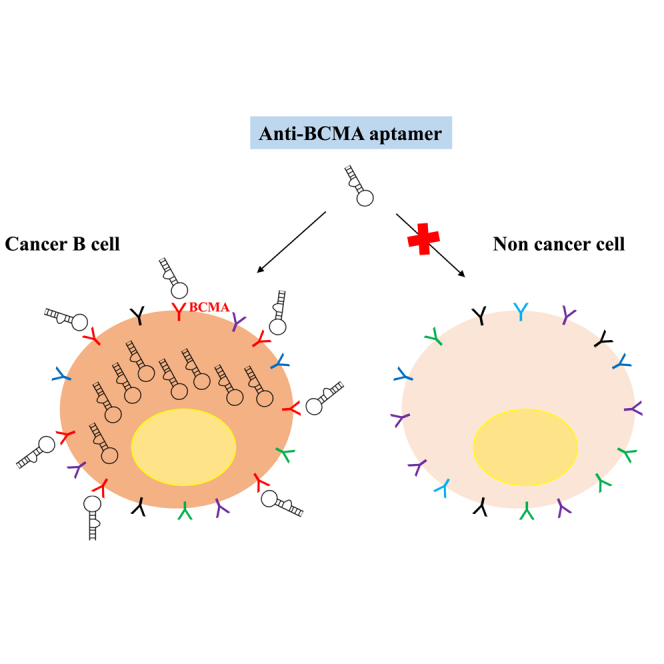
B cell maturation antigen is highly expressed on malignant plasma cells in human multiple myeloma and has recently emerged as a very promising target for therapeutic interventions. Nucleic-acid-based aptamers are small oligonucleotides with high selective targeting properties and functional advantages over monoclonal antibodies, as both diagnostic and therapeutic tools. Here, we describe the generation of the first-ever-described nuclease resistant RNA aptamer selectively binding to B cell maturation antigen. We adopted a modified cell-based systematic evolution of ligands by exponential enrichment approach allowing the enrichment for internalizing aptamers. The selected 2′Fluoro-Pyrimidine modified aptamer, named apt69.T, effectively and selectively bound B cell maturation antigen-expressing myeloma cells with rapid and efficient internalization. Interestingly, apt69.T inhibited APRIL-dependent nuclear factor κB (NF-κB) pathway in vitro. Moreover, the aptamer was conjugated to microRNA-137 (miR-137) and anti-miR-222, demonstrating high potential against tumor cells. In conclusion, apt69.T is a novel tool suitable for direct targeting and delivery of therapeutics to B cell maturation antigen-expressing myeloma cells.
Keywords: aptamers, multiple myeloma, BCMA, targeted delivery, cell-internalizing SELEX, aptamer-based conjugates
Graphical Abstract
Introduction
Multiple myeloma (MM) is an aggressive hematologic disease that accounts for 1% of all cancers and about 13% of all hematologic malignancies. It is characterized by an abnormal proliferation and accumulation of malignant immunoglobulin-producing plasma cells (PCs) in the bone marrow (BM), associated with systemic signs including osteolytic bone lesions. Despite considerable recent advances in MM therapy that have significantly improved the overall survival of patients, it still remains in most cases an incurable lethal disease. Indeed, currently available standard-of-care therapies for MM patients, which include autologous or allogeneic stem cell transplantation, innovative immunomodulatory agents, CD38- and SLAMF7-directed monoclonal antibodies (mAbs), proteasome inhibitors, and dendritic cell-based vaccines, show effectiveness in a limited subset of patients.1,2 Thus, new effective treatments are urgently awaited to improve patient clinical outcome.
In recent years, among several new investigational therapeutic strategies toward new targets,3, 4, 5, 6 B cell maturation antigen (BCMA) has emerged as promising for immunotherapy against MM. Also known as tumor necrosis factor receptor superfamily member 17 (TNFRSF17), BCMA is a member of the TNF receptor superfamily and is functionally related to the transmembrane activator and calcium modulator and cyclophilin ligand (TACI or TNFRSF13B) and B cell activating factor receptor (BAFF-R or TNFRSF13C) receptors. BCMA is found on the cell surface of late-stage normal B lymphocytes and highly expressed on malignant PCs from MM patients.7, 8, 9 The binding of its specific ligands, BAFF and the proliferation-inducing ligand (APRIL or TNFSF13), induces activation of canonical and non-canonical nuclear factor κB (NF-κB) pathway and promotes long-lived PCs survival.10,11 Recently, it has been demonstrated that BCMA also exists in a soluble form.12 Different evidences showed that serum BCMA (sBCMA) is a promising marker for monitoring the course of the disease in MM patients and that it does not represent an obstacle to the success of the anti-BCMA-targeted therapies.13
On the basis of these considerations, mAbs targeting BCMA have been developed as tools for MM-targeted therapy. These mAbs inhibit APRIL-dependent activation of downstream NF-κB and support immune-mediated killing of tumor cells.14,15 To improve the antitumor potential of approaches targeting BCMA, a mAb has been modified by conjugation to cytotoxic drugs, such as monomethylauristatin F, a potent inhibitor of tubulin polymerization.16,17 Moreover, BCMA/CD3 bispecific mAbs and T cells genetically engineered to express chimeric antigen receptors (CAR-Ts) targeting BCMA have demonstrated significant anti-MM activity and are currently in early clinical trials.7,17, 18, 19, 20, 21, 22, 23
In last decades, nucleic acid aptamers have emerged as effective therapeutics for a wide range of human pathologies, including solid and hematological tumors.24 Aptamers are also called “nucleic acid monoclonal antibodies” due to their capacity to fold in particular three-dimensional shapes and to bind specific targets. They show many advantages over monoclonal antibodies as diagnostic and therapeutic tools, such as low immunogenicity, small size, high batch fidelity, easy production, increased chemical stability, and versatility. Interestingly, aptamers directed against cell surface receptors represent very attractive options for therapeutic targeted delivery.25,26 Indeed, upon binding, aptamers undergo intracellular uptake mediated by the target receptor and have been used to drive therapeutic molecules selectively to cells overexpressing the specific target.
In this work, we address the generation and the characterization of a 2′Fluoro-Pyrimidine (2′F-Py) RNA aptamer, named apt69.T, selectively binding BCMA. We provide here proof-of-concept data that support the apt69.T anti-BCMA aptamer as potential new tool for the experimental treatment of MM.
Results
Generation of Internalizing RNA Aptamers Targeting BCMA
In order to select aptamers specific for binding to BCMA receptor, and able to internalize in a receptor-mediated manner as well, we adopted a variant of the conventional cell-SELEX (systematic evolution of ligands by exponential enrichment)27 procedure (see Table S1), introducing in the last three rounds a partitioning step of high-salt-resistant aptamers thus enriching for aptamers capable of rapid and selective cell internalization (cell-internalizing SELEX).28,29
As shown in Figure S1A, in the counter-selection step, a starting 2′F-Py RNA aptamer library pool was incubated with parental COS-7 BCMA-negative cells, and then, in the positive selection step, unbound aptamers were recovered and incubated with the same cell line transiently transfected with BCMA receptor (COS-7-BCMA). Overexpression of BCMA in COS-7 parental cells upon transfection was confirmed by western blot analysis (Figure S1B). Following 11 rounds of cell-SELEX (rounds 1–8 followed by intracellular rounds 9–11), the final aptamer pool was cloned and 11 aptamer sequences, each present in two or three copies in the alignment (Figure S1C), were validated for the binding activity on BCMA-expressing cells.
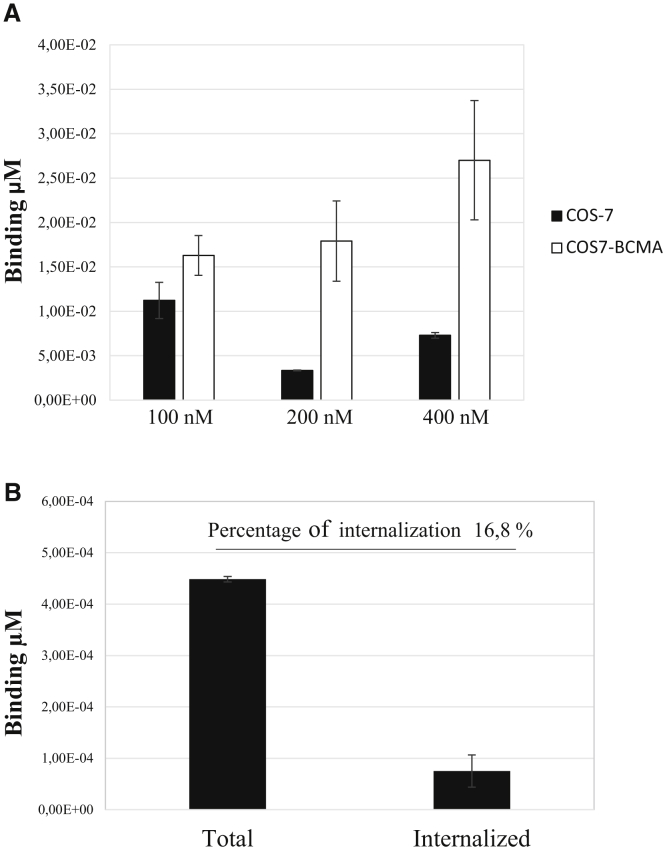
As assessed by qRT-PCR-based binding assay, among the 11 analyzed aptamers, the apt69 and the apt29 showed the capacity to discriminate COS-7-BCMA cells from parental COS-7 cells at 100 nM concentration (Figures S2A and S2B). Further investigations revealed that apt69 holds the best binding ability and is able to effectively discriminate COS-7-BCMA cells from parental COS-7 cells in a dose-dependent analysis (Figure 1A). Specificity of the binding was also confirmed comparing apt69 binding ability with that of an unrelated control aptamer (UnrelApt) on COS-7-BCMA cells (see Figure S3). Further, the ability of the apt69 to internalize into BCMA expressing target cells has been evaluated by qRT-PCR to be ~16.8% of the total aptamer bound (Figure 1B). Based on the obtained results, apt69 was chosen for further characterization.
Figure 1.
Apt69 Binding and Internalization
(A) Apt69 was incubated at the indicated concentrations with COS-7 parental cells or with COS7-BCMA cells for 30 min at 37°C. Binding values (μM) were measured by qRT-PCR. (B) COS7-BCMA cells were incubated for 1 h with apt69 (200 nM) and either analyzed (total) or subjected to high salt washing before harvesting (internalized). The quantities of total and internalized aptamer were evaluated by qRT-PCR. Error bars show the mean ± SEM values.
Aptamer Truncation
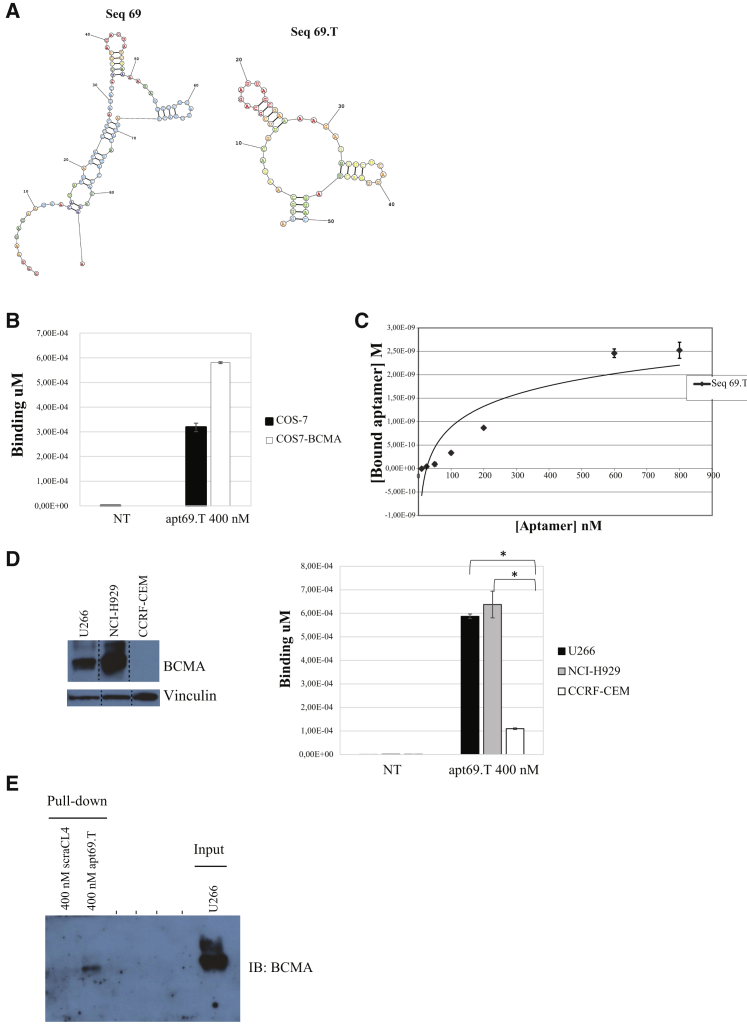
We thus proceeded by reducing apt69 to a length compatible with chemical synthesis. Based on bioinformatic prediction of its secondary structure (RNAstructure, https://rna.urmc.rochester.edu and RNAfold, http://rna.tbi.univie.ac.at), we designed a shorter apt69 version in which the two central stem loops were preserved while the entire 5′ and part of the 3′-constant regions were removed (Figure 2A). The resulting 50-mer aptamer, named apt69.T, was then validated in vitro for target binding.
Figure 2.
Apt69.T Specificity
(A) Secondary structure of apt69 full-length aptamer (left) and apt69.T truncated version (right) predicted by RNAstructure Fold. (B) Binding of apt69.T on COS-7 parental cells and COS7-BCMA. Cells were incubated with 200 nM apt69.T for 30 min at 37°C. Binding values (μM) were evaluated by qRT-PCR. Error bars show the mean ± SEM values (C) Determination of the dissociation constant (KD) of apt69.T on COS7-BCMA cells. Parental COS-7 and COS7-BCMA cells were incubated at 37°C with increasing concentrations of the aptamer. The binding data obtained by qRT-PCR were fitted, and KD was calculated. In all binding assays, the background values obtained with an unrelated aptamer were subtracted from the values obtained with the apt69.T. Error bars show the mean ± SEM values. (D) Apt69.T (400 nM) was incubated for 15 min with U266 (BCMA+), NCI-H929 (BCMA+), and CCRF-CEM (BCMA−) cells. Binding values (μM) were evaluated by qRT-PCR. Error bars show the mean ± SEM values. Statistics were calculated using Student’s t test, *p < 0.05. (E) Aptamer-mediated pull-down. U266 cells were incubated with the indicated concentration of biotinylated apt69.T or a control aptamer (scraCL4). Cell lysates were purified on streptavidin beads and immunoblotted with anti-BCMA antibody. 5 μg of total cell extracts (Input) were loaded as a reference.
As assessed by qRT-PCR-based binding assay, the shorter aptamer apt69.T preserves the ability to selectively bind to COS-7-BCMA cells and efficiently discriminates them from parental COS-7 cells (Figure 2B) with an apparent dissociation constant (KD = 79.4 nM) (Figure 2C). Further, we also determined the BCMA-dependent binding of the apt69.T to human malignant myeloma cells. As shown, the apt69.T revealed that it preferentially binds human B lymphocytes from MM, U266, and NCI-H929 BCMA-expressing cells, as compared with T lymphoblastoid cell line (CCRF-CEM) acute lymphoblastic leukemia T lymphoblasts that do not express detectable levels of BCMA (Figure 2D, left and right panels). On the other hand, the CCRF-CEM cells express TACI30 but neither BCMA nor BAFF-R,31 while U266 (BCMA+) cells express TACI, but not BAFF-R;32 and NCI-H929 (BCMA+) cells do not express TACI and show a moderate expression of BAFF-R.15 Results shown well support the binding of apt69.T to BCMA expressed on the cell surface of MM cells, but not, or poorly, to the functional related receptors for either APRIL and BAFF or TACI and BAFF-R. Finally, we confirmed binding of the apt69.T to BCMA by performing an aptamer-mediated affinity pull-down assay. To this end, MM U266 BCMA-expressing cells were treated with biotin-tagged apt69.T and total cell extracts purified on streptavidin-coated beads, followed by immunoblotting with anti-BCMA antibody. As shown in Figure 2E, the apt69.T is able to interact with BCMA, whereas no binding was obtained with an unrelated 2′F-Py RNA aptamer used as a negative control.
Taken together, these data indicate that the short version of apt69, the apt 69.T, is a highly specific affinity ligand for the BCMA receptor.
Apt69.T Serum Stability and Functional Activity
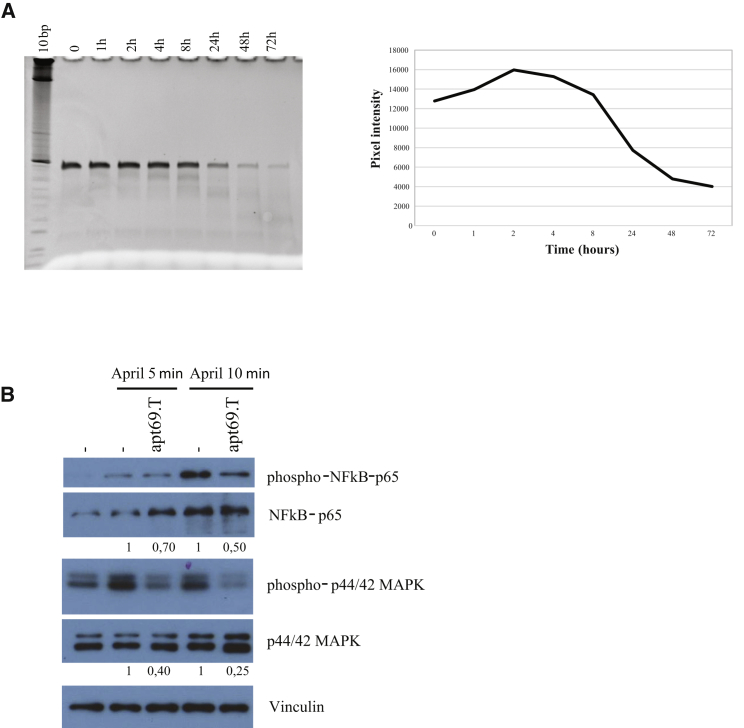
An important obstacle to the development of RNA-based drugs is represented by their rapid clearance and poor stability in the circulation. Indeed, RNA is highly sensitive to the degradation by the presence of nucleases in serum. Apt69.T contains 2′F-Py modifications that confer to the aptamer increased resistance to nuclease degradation.33 In order to evaluate apt69.T serum stability, we incubated the aptamer in 80% human serum at 37°C for increasing times, up to 72 h. Then, the integrity of RNA samples was analyzed by denaturing PAGE (Figure 3A, left panel). As shown, the aptamer remained almost stable up to 8 h and then gradually degraded. Then, in order to calculate the approximate half-life, band intensities have been quantified by using ImageJ program (Figure 3A, right panel). The estimated half-life for the apt69.T in 80% human serum is calculated to be ~20 h. Further, in order to investigate whether the apt69.T binding causes functional inhibition of the ligand-dependent BCMA activity, we evaluated the APRIL-dependent activation of NF-κB pathway upon aptamer treatments. In order to exclude any interference from the TACI receptor, of which APRIL is a shared ligand, we decided to perform this analysis on NCI-H929 B cells that express a high level of BCMA but do not express TACI.30 Starved NCI-H929 cells were treated for 30 min with 400 nM of apt69.T and then stimulated with human APRIL ligand for 5 or 10 min in the presence or not of the aptamer. We noticed that the apt69.T is able to reduce APRIL-induced phosphorylation of p65 reaching a reduction of ~50% following 10 min of stimulation. We also observed a clear inhibition of downstream effectors extracellular signal-regulated kinase (ERK) 1/2 phosphorylation (Figure 3B), up to 75% at 10 min of APRIL stimulation.
Figure 3.
In Vitro Aptamer Activity
(A) Apt69.T serum stability was measured by incubating the aptamer at 4 μM in 80% human serum for indicated times. At each time point, RNA-serum samples were collected and evaluated by electrophoresis with 15% denaturing polyacrylamide gel. Gel was stained with ethidium bromide and visualized at the Gel Doc EZ Imager (left panel); bands were quantified by using ImageJ program and pixel intensity for each time point is reported in the graph (right panel). (B) To evaluate apt69.T inhibition of APRIL-induced NF-κB canonical pathway, we supplemented NCI-H929 cells starved with RPMI medium with 0.25% FBS for 24 h. Cells were treated with apt69.T (400 nM) for 30 min at 37°C and then stimulated with human recombinant APRIL ligand (400 ng/mL) for indicated times in the presence or not of the aptamer. Total cell lysates (4 μg) were immunoblotted with anti-phospho-p44/42 MAPK (Thr202/Tyr204), anti-phospho-NF-κB-P65 (Ser536), anti-ERK 1, anti-NF-κB-P65, and anti-vinculin antibodies.
Taken together, these data indicate that the apt69.T is able to inhibit BCMA downstream pathway.
Apt69.T Internalization into BCMA-Expressing Cells and Targeted Delivery of miRNAs and miRNA Antagonists
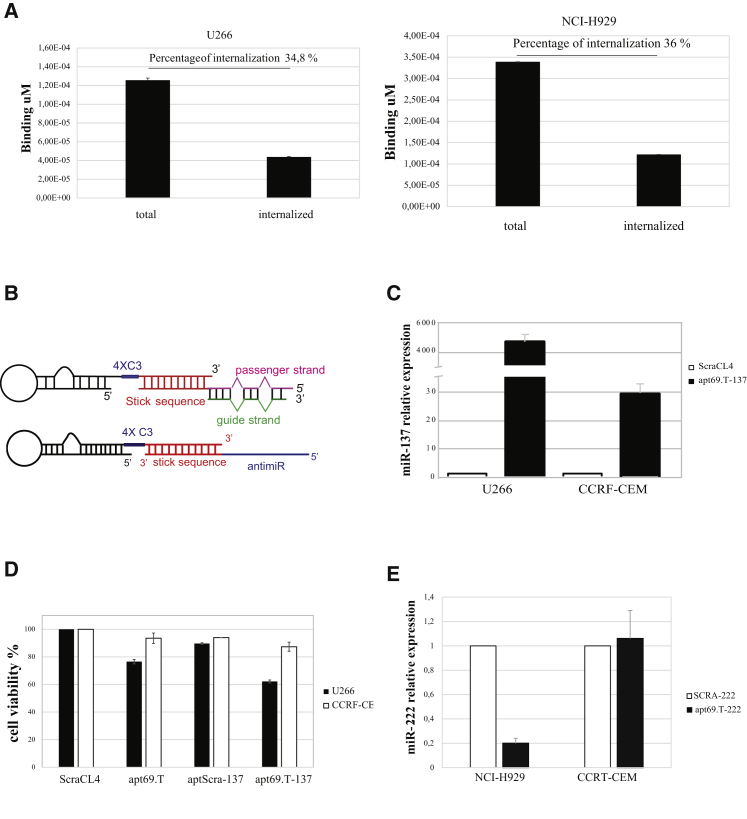
In order to verify that the truncation process does not alter the capacity of the aptamer to internalize into target cells, we then analyzed the in vitro aptamer uptake in BCMA-expressing MM cells. As assessed by binding for high-salt-resistant aptamer, we observed that apt69.T rapidly internalizes into U266 and H929 cells, showing approximately 35% of internalization following 15 min treatment in both cell lines (Figure 4A, left and right panel). Aptamer internalization was further increased following longer incubation times, reaching to ~50% following 2 h of exposition (Figure S4). Taking into account that the apt69.T half-life is of about 20 h (Figure 3A), apt69.T may represent a promising carrier for targeted therapy in MM.
Figure 4.
Apt69.T Aptamer Internalization and miRNA/anti-miR Delivery In Vitro
(A) Internalization of apt69.T on U266 (left) and H929 (right) (BCMA+) cells. Apt69.T (200 nM) was incubated for 15 min with cells and then cells were recovered following three washings with PBS (total) or with PBS 0.5 M NaCl (internalized) in order to remove aptamer on the cell surface. The quantities of total and internalized aptamer were evaluated by qRT-PCR. Error bars show the mean ± SEM values. (B) Schematic representation of the apt69.T-miR-137 conjugate (top) and apt69.T-anti-miR-222 (bottom). (C) U266 (BCMA+) or CCRF-CEM (BCMA−) cells were treated (400 nM) with indicated molecules. miR-137 level relative to cells treated with control aptamer (scraCL4) was evaluated by qRT-PCR following 48 h. Bars show the mean ± SD values. (D) U266 (BCMA+) and CCRF-CEM (BCMA−) cells were treated (400 nM) with indicated molecules. 72 h post-treatment, cell viability was measured by MTT assay. Error bars show the mean ± SD values. (E) NCI-H929 (BCMA+) or CCRF-CEM (BCMA−) cells were treated (400 nM) with indicated molecules. miR-222 level relative to cells treated with control conjugate (SCRA-222) was evaluated by qRT-PCR following 72 h. Error bars show the mean ± SD values.
Emerging evidence demonstrates that aptamers, thanks to their advantageous properties over monoclonal antibodies, are ideal carrier molecules for selective delivery of a potential huge variety of therapeutics. As proof-of-concept study, we studied the potential delivery of non-coding RNAs (ncRNAs) or synthetic oligonucleotides (ONs), such as small interfering RNAs (siRNAs), microRNAs (miRNAs), and miRNA antagonists that have been widely demonstrated to exert anti-tumor activity in different tumors, including MM.4,25,34, 35, 36, 37, 38, 39, 40, 41 As shown in Figure 4B, by using a stick-end based approach, previously described for the targeted delivery of RNA therapeutics,25,36,42 we designed two different conjugates. In the first, the passenger strand of the mature human miR-137 was conjugated with the 3′ end of the apt69.T, reported to be a tumor suppressor miRNA in MM.43,44 In the second, the apt69.T was instead conjugated to a single strand antagonist of miR-222, an oncogenic miRNA found overexpressed in MM.45 U266 cells, expressing basal low levels of miR-137, comparable to that expressed by CCRF-CEM, were treated with aptamer 69.T-miR-137 stick conjugate (indicated as apt69.T-137). The apt69.T-137 conjugate demonstrated that it induces a strong upregulation of the miR-137 in BCMA-expressing-U266 cells. A minimal increase of cellular miR-137 expression is also observed in CCRF-CEM cells, probably due to a slight aspecific cell uptake. However, induced miR-137 expression level was ~170-fold lower than that observed in BCMA-expressing U266 cells (Figure 4C). We then explored the effect of apt69.T-137 conjugate on myeloma cell growth and observed a clear reduction (~40%) of cell viability in BCMA-expressing-U266 cells, following 72 h treatment with 400 nM apt69.T-137 conjugate (Figure 4D).
Further, NCI-H929 BCMA-expressing cells and CCRF-CEM control cells, showing similar measurable miR-222 basal levels, were treated with the apt69.T-anti-miR-222 stick conjugate (indicated as apt69.T-222), and miR-222 levels were measured by qRT-PCR. An evident downregulation of miR-222 expression levels (~80%) was observed in NCI-H929 cells, while no detectable effect was observed in CCRF-CEM BCMA-negative cells (Figure 4E). Collectively, these data indicate that apt69.T can be developed as an efficient and specific delivery carrier of therapeutic RNAs.
Discussion
MM is an incurable malignancy and much effort has been devoted over the years in the translational research for the identification of novel agents specifically targeting tumor cells, including the use of nucleic-acid-based agents.37,39,46, 47, 48, 49, 50, 51 However, differently from other nucleic acid-based molecules, aptamers are emerging as innovative potential therapeutic tools due to their three-dimensional shapes that confer monoclonal antibody-like properties for the binding to specific cell surface antigens.
Here we reported the generation of a novel unique 2′Fluoro-Pyrimidine modified aptamer directed against BCMA. The aptamer we selected, named apt69.T, resulted to hold excellent serum stability with an estimated half-life of ~20 h in 80% human serum and showed effective binding to human BCMA on MM cells in vitro (Figures 2B–2E and 3A). Interestingly, the apt69.T inhibited APRIL-dependent BCMA downstream NF-κB pathway (Figure 3B) and was rapidly internalized into target-expressing cells, providing therefore an effective tool for delivery of therapeutics, as demonstrated by miRNAs and miRNA antagonists delivery (Figures 4A–4E).
To our knowledge, our aptamer is the first-ever-described aptamer-targeting BCMA. This antigen is a well-recognized target whose expression is highly restricted to terminally differentiated B cells and overexpressed in malignant MM cells, where it plays a role for cell survival. Monoclonal antibodies and CAR-Ts directed against BCMA have been developed and validated both in vitro and in vivo, providing a proof of principle that targeting BCMA is a reliable approach for MM treatment.14, 15, 16, 17,20,22,52, 53, 54 However, their clinical applications have been associated with some difficulties and safety issues.55 In this light, nucleic acid aptamers show recognition and affinity properties similar to those of mAbs but are characterized by a cheaper and cost-effective production and have a potentially very low toxicity. Even if aptamer toxicity or immunogenicity has not yet been deeply explored, it is well known that antibodies against synthetic oligonucleotides are not generally produced and that the Toll-like receptor-mediated innate immune response, commonly activated by non-self nucleic acids, is abrogated by 2′-modified nucleotides containing aptamers.56,57 These observations suggest a safety profile of aptamers that warrants further investigation.
To date, an increasing number of aptamers provide innovative tools for cancer therapy. In some cases, they can be used as inhibitory molecules, and in other cases, thanks to their excellent targeting and internalization properties, they can be used as carriers for secondary therapeutic agents.
The apt69.T we selected has been isolated by using a protocol adapted from Giangrande PH group to select internalizing aptamers in non-adherent cell lines.25 The aptamer demonstrated the capacity to bind with a good affinity and specificity BCMA on MM cells in vitro. Moreover, the apt69.T seems to be a very interesting molecule because it seems to hold both inhibitory and carrier capabilities. Indeed, it revealed the ability to inhibit APRIL-induced NF-κB pathway and also to effectively deliver double-strand miRNA and single-strand miRNA antagonist molecules in BCMA-expressing MM cells. Taken together, our findings indicate that the apt69.T is a very promising tool for MM-targeted therapy, providing therefore proof-of-concept evidence and a framework for the future development of novel and effective aptamer-based targeted therapies for MM patients.
Materials and Methods
Cell Cultures and Transfection
Cell lines were purchased from the ATCC (LG Standards, Milan, Italy). COS-7 cells were grown in DMEM, whereas human U266, NCI-H929, and CCRF-CEM cells were grown in RPMI. Media were supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin. All cell culture reagents were purchased from Sigma (St. Louis, MO, USA).
The day before transfection, cells were seeded in 10% FBS. All transfections were performed using the serum-free Opti-MEM and Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. BCMA plasmid was provided by OriGene Technologies (9620 Medical Center Drive, Suite 200, Rockville, MD 20850, USA).
Cell-SELEX Procedure
The starting aptamer pool used in the SELEX procedure was composed by RNA sequences modified with 2′F-Py (TriLink BioTechnologies, San Diego, CA, USA) containing a central 40 bp-long randomized region and two 23 bp-long fixed regions for RT-PCR. The forward selection primer (FSP) sequence was: 5′-TAGGGAAGAGAAGGACATATGAT-3′; the reverse selection primer (RSP) was: 5′-TCAAGTGGTCATGTACTAGTCAA-3′. Parental COS-7 (BCMA−) cells were used in the counter-selection step, while COS-7 transiently transfected with human BCMA were used in the positive selection step (COS-7-BCMA). The RNA pool was first incubated with COS-7 cells (counter-selection step), and then unbound aptamers were incubated with COS-7-BCMA cells (selection step). Bound aptamers were recovered and RT-PCR and in vitro transcription was performed. After eight rounds of selection and counter-selection, three additional rounds of selection were performed, introducing a step of high salt washings with Dulbecco’s phosphate-buffered saline (DPBS) 0.5 M NaCl downstream of the selection step, in order to remove aptamers on the cell surface not able to internalize into target cells. The enrichment for nonspecific ligands has been avoided by modulating the stringency at each SELEX round in different ways; for example, by using poliinosinic acid as an unspecific competitor during the incubation step, reducing the target concentration, increasing the number of counter-selection steps, and increasing the number of washings after selection. Following a total of 11 SELEX rounds, the resulting pool has been cloned and ~100 individual clones were sequenced.
Bioinformatics Analysis
Single aptamer sequences obtained were analyzed by using Multiple Sequence Alignment tool (ClustalW2) to identify aptamer sequence similarities.
Aptamer secondary structures were predicted by using RNAstructure Fold algorithms.
Aptamer Sequences
apt29:
5′-GGGAAGAGAAGGACAUAUGAUCCUAGUCUUGAAUGCACCUCGGGUCAGAAGGGGUUCCAGCUUGACUAGUACAUGACCACUUGA-3′
apt69:
5′-GGGAAGAGAAGGACAUAUGAUCAGUGCAAGACGUUCGCAGAUUAGCGAAAAGAGGGUCUCAUUGACUAGUACAUGACCACUUGA-3′
apt69.T:
5′-AGUGCAAGACGUUCGCAGAUUAGCGAAAAGAGGGUCUCAUUGACUAGUAC-3′
apt69.T (Biotin):
5′-AGUGCAAGACGUUCGCAGAUUAGCGAAAAGAGGGUCUCAUUGACUAGUAC{BioBB}-3′
Scrambled CL4 (scraCL4):
5′-UUCGUACCGGGUAGGUUGGCUUGCACAUAGAACGUGUCA-3′
scraCL4 (Biotin):
5′-UUCGUACCGGGUAGGUUGGCUUGCACAUAGAACGUGUCA{BioBB}-3’
Unrelated Control Aptamer (Indicated as UnrelApt):
5′-GGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCU-3′
apt69.T Stick:
5′-AGUGCAAGACGUUCGCAGAUUAGCGAAAAGAGGGUCUCAUUGACUAGUACXXXXGUACAUUCUAGAUAGCC-3′
SCRA Stick (Indicated as SCRA):
5′-GGCGCUAGAACCUUCUAAGCGAAUACAUUACCGCXXXXGUACAUUCUAGAUAGCC-3′
miR-137 Passenger Stick:
5′-GGUGACGGGUAUUCUUGGGUGGAUAAUAGGCUAUCUAGAAUGUAC-3′
miR-137 Guide:
5′-UGUUAUUGCUUAAGAAUACGCGUAGUCUU-3′
anti-miR-222 Stick:
5′ ACCCAGUAGCCAGAUGUAGCUGGCUAUCUAGAAUGUAC 3′;
Scra Aptamer (Indicated as aptScra):
5′-GCUCAACGUUGACAACAAAUGACGAXXXXGUACAUUCUAGAUAGCC-3′
All RNA sequences contain 2′F-Py. Long aptamers (apt29 and apt69) were produced by in vitro transcription using the correspondent DNA as template in the presence of 2′F-Py (tebu-bio SRL, Magenta, Milan, Italy). apt69.T, apt69.T stick, miR-137 passenger stick, and miR-137 guide were synthesized by TriLink Bio Technologies (San Diego, CA, USA) or Synthetic and Biopolymer Chemistry Core at the Beckman Research Institute of City of Hope (Duarte, CA, USA). Stick sequences (underlined) consist of 2′F-Py and 2′-oxygen-methyl purines. The italic X indicates a covalent spacer, a three-carbon linker ([CH2]3).
Before treating cells, aptamers were subjected to a short denaturation-renaturation step by incubating 5 min at 85°C, 5 min on ice, and 10 min at 37°C.
Binding Assays by qRT-PCR
24 h following transient transfection of COS-7 cells with human BCMA, parental COS-7 cells and COS-7-BCMA cells were seeded in 6-well plates (1.6 × 105 cells/well). The next day, cells were treated with the indicated concentrations of the aptamer for 30 min at 37°C in serum-free medium.
For binding on U266, NCI-H929, and CCRF-CEM, 4 × 105 cells/well were pretreated for 30 min at 37°C with 0.4 mg/mL tRNA and then treated with 200 nM aptamer for 15 min at 37°C in serum-free medium. Unbound RNA was removed by washing 3 times with ice-cold DPBS. Bound RNA was recovered by TRIzol containing 0.5 pmol/mL of CL4 aptamer (CL4: 5′-GCCUUAGUAACGUGCUUUGAUGUCGAUUCGACAGGAGGC-3′) used as a reference control.
The amount of recovered RNA was determined by performing a two-step qRT-PCR protocol. In step one, recovered RNA was reverse transcribed using an RNA-dependent DNA polymerase encoded by Moloney Murine Leukemia Virus (MMuLV) and specific 3′ primers. The primers used for the RT step were as follows: RSP (for both apt29 and apt69) 5′-TCAAGTGGTCATGTACTAGTCAA-3′; primer apt69.T Rv 5′-GTACTAGTCAATGAGACCCT-3′; primer CL4 Rv 5′-GCCTCCTGTCGAATCG-3′.
The protocol used for RT was as follows: heating step at 65°C for 5 min, annealing step at 22°C for 5 min, extension at 42°C for 30 min followed by end extension at 48°C for 30 min and enzyme inactivation at 95°C for 5 min. In step two, the products from the RT reaction were PCR amplified with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). The qPCR protocol used was: heating of the RT products at 95°C for 2 min, followed by 40 cycles of heating at 95°C for 30 s, annealing at 55°C for 30 s, extending at 60°C for 30 s. A melt curve stage by heating at 60°C–95°C was performed. Primers used for the qPCR were as follows: FSP Seq 5′-TAGGGAAGAGAAGGACATATGAT-3′; RSP 5′-TCAAGTGGTCATGTACTAGTCAA-3′; primer apt69.T Fw 5′-AGTGCAAGACGTTCGCAGA-3′; primer apt69.T Rv 5′-GTACTAGTCAATGAGACCCT-3′; primer CL4 Fw 5′-GCCTTAGTAACGTGCTTT-3′ primer CL4 Rv 5′-GCCTCCTGTCGAATCG-3′.
Data were normalized to the CL4 reference control and to cell number, as determined by counting cells cultured in conjunction with each experiment.
For binding-affinity evaluation, parental COS-7 or COS-7-BCMA cells were incubated with the indicated concentrations of the apt69.T aptamer for 30 min at 37°C in serum-free medium. Lineweaver-Burk analysis was performed for the apparent binding constant (KD) calculation.
Internalization Assays by qRT-PCR
BCMA+ cells (COS-7-BCMA, U266, or H929) were incubated for indicated times with 200 nM aptamer in serum-free medium. After treatment, cells were washed DPBS or with DPBS 0.5 M NaCl in order remove not internalized aptamers. Binding was evaluated by qRT-PCR and the percentage of internalized aptamers on the total bound was calculated.
Aptamer Stability in Human Serum
apt69.T aptamer was incubated 4 μM in 80% human serum from 1 h to 72 h. Type AB Human Serum provided by Euroclone (category ECS0219D) was used. At each time point 4 μL (16 pmol RNA) was withdrawn and incubated for 1 h at 37°C with 5 μL of proteinase K solution (600 mAU/mL) in order to remove serum proteins that interfere with electrophoretic migration. Following proteinase K treatment, 18 μL denaturing RNA dye (Invitrogen, Waltham, MA, USA) were added to samples that were then stored at −80°C. All time point samples were separated by electrophoresis into 15% denaturing polyacrylamide gel. The gel was stained with ethidium bromide and visualized by UV exposure.
Western Blot Analysis
To evaluate aptamer inhibition of APRIL-induced NF-κB signaling, we seeded NCI-H929 cells in 6-well plates (3 × 105 cells/well) and starved them with 0.25% FBS RPMI medium for 24 h. Cells were treated with apt69.T (400 nM) for 30 min at 37°C and then stimulated with 400 ng/mL of human recombinant APRIL ligand (R&D Systems, Minneapolis, MN, USA) for 5 and 10 min in the presence or not of the apt69.T. Total cell lysates were prepared in JS buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 5 mM EGTA, 1 mM Na3VO4, protease inhibitors) and then boiled in SDS/β-mercaptoethanol sample buffer. Proteins were separated by electrophoresis and then blotted onto a nitrocellulose membrane (GE Healthcare, Chicago, IL, USA) by an electrophoretic transfer. After blocking with 5% BSA in Tris-buffered saline (TBS) containing 0.1% Tween-20, membranes were incubated at 4°C overnight with the following primary antibodies: anti-phospho-p44/42 mitogen-activated protein kinase (MAPK) (Thr202/Tyr204), anti-vinculin (Cell Signaling Technology, Danvers, MA, USA); anti-phospho-NF-κB-P65 (Ser536), anti-NF-κB-P65 (Elabscience Biotechnology, Houston, TX, USA); anti-ERK 1 (Santa Cruz Biotechnology, Stockton, CA, USA).
Aptamer-Mediated Pull-Down
1 × 106 U266 cells were incubated in serum-free medium at 37°C for 30 min with 400 nM of apt69.T biotinylated at the 3′ end or with similarly biotinylated scraCL4 sequence, used as a control. Following three washings with DPBS, cells were lysed with 10 mmol/L Tris-HCl pH 7.5 containing 200 mmol/L NaCl, 5 mmol/L EDTA, 0.1% Triton X-100, and protease inhibitors.
Cell extracts (800 μg in 0.8 mL lysis buffer) were purified by incubating with 480 μL streptavidin beads (ThermoFisher Scientific) for 2 h with rotation. Following incubation, beads were washed three times with DBPS and bound proteins were recovered by adding Laemmli buffer and then analyzed by immunoblotting with anti-BCMA antibody (Novus Biologicals, CO, USA).
Aptamer-miRNA/anti-miR Conjugate Production
To prepare aptamer-miRNA conjugates, we annealed the miR-137 passenger and guide by incubating in annealing buffer (20 mM 2-[4-(2- hydroxyethyl) piperazin-1-yl] ethane sulfonic acid [HEPES] pH 7.5, 150 mM NaCl, 2 mM CaCl2) for 10 min at 95°C, 10 min at 55°C, and then 20 min at 37°C. The stick aptamer was refolded (5 min 85°C, 5 min on ice, 10 min at 37°C) and then equal amounts of stick aptamer and annealed passenger-guide strands were incubated at 37°C for 30 min to allow conjugation through the complementary stick sequences.
To prepare aptamer-anti-miR conjugates, we refolded the stick aptamer as indicated above and incubated the anti-miR-stick at 95°C for 10 min in the annealing buffer. Then, an equal amount (ratio 1:1) of stick aptamer and anti-miR-stick was annealed by incubation at 37°C for 30 min.
The annealing efficiency was evaluated on a 12% non-denaturing polyacrylamide gel.
miRNA and anti-miR Delivery Assays
1.4 × 105 cells were seeded in 6-well plates and treated with 400 nM of apt69.T-137 (apt69.T-miR-137 conjugate), apt69.T-222 (apt69.T-anti-miR-222 conjugate), control chimera (SCRA-222), or control aptamer (scraCL4) for 48 or 72 h. Cells were then washed with ice-cold DPBS and total RNA was extracted with Total RNA Purification Plus Kit (Norgen Biotek,ON, Canada). 500 ng of total RNA was then reverse transcribed with miScript Reverse Transcription kit (QIAGEN, Milan, Italy), according to the manufacturer protocol. Amplification, to evaluate miRNA expression level, was performed by using the miScript-SYBR Green PCR Kit and specific miScript Primer Assay (QIAGEN, Milan, Italy). U6 RNA was used as a housekeeping control gene.
Cell Viability
7 × 103 cells were seeded in 96-well plates and treated with 400 nM of scraCL4, apt69.T, aptScra-137 (scrambled stick aptamer conjugated with miR-137), or apt69.T-137. Cell viability was measured by methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay (category number M 5655, Sigma, St. Louis, MO, USA), according to manufacturer instructions.
Author Contributions
S.C. designed and performed the majority of the experiments, interpreted results, and guided manuscript preparation. M.T.D.M. and C.L.E. performed part of experiments and assisted with manuscript preparation. S.N. assisted with some experiments. V.d.F. and P.T. provided intellectual input, coordinated the research, secured the funding, guided result interpretation, and supervised manuscript preparation.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We wish to thank D. Rotoli, L. Baraldi, and F. Moscato for technical assistance. This work has been supported by the Italian Association for Cancer Research (AIRC) with “Special Program for Molecular Clinical Oncology–5 per mille,” 2010/15 and its Extension Program” number 9980, 2016/18 to P.T. and V.d.F.; “Innovative Immunotherapeutic Treatments of Human Cancer” multi-unit regional number 16695 (cofinanced by AIRC and the CARICAL Foundation), and the Italian Ministry of Health (GR-2011-02352546; C.L.E.). S.C. was supported by POR CAMPANIA FSE 2014/2020.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.021.
Contributor Information
Pierfrancesco Tassone, Email: tassone@unicz.it.
Vittorio de Franciscis, Email: defranci@unina.it.
Supplemental Information
References
- 1.Kumar S. Emerging options in multiple myeloma: targeted, immune, and epigenetic therapies. Hematology (Am. Soc. Hematol. Educ. Program) 2017;2017:518–524. doi: 10.1182/asheducation-2017.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larocca A., Mina R., Gay F., Bringhen S., Boccadoro M. Emerging drugs and combinations to treat multiple myeloma. Oncotarget. 2017;8:60656–60672. doi: 10.18632/oncotarget.19269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi M., Di Martino M.T., Morelli E., Leotta M., Rizzo A., Grimaldi A., Misso G., Tassone P., Caraglia M. Molecular targets for the treatment of multiple myeloma. Curr. Cancer Drug Targets. 2012;12:757–767. doi: 10.2174/156800912802429300. [DOI] [PubMed] [Google Scholar]
- 4.Amodio N., Di Martino M.T., Neri A., Tagliaferri P., Tassone P. Non-coding RNA: a novel opportunity for the personalized treatment of multiple myeloma. Expert Opin. Biol. Ther. 2013;13(Suppl 1):S125–S137. doi: 10.1517/14712598.2013.796356. [DOI] [PubMed] [Google Scholar]
- 5.Botta C., Gullà A., Correale P., Tagliaferri P., Tassone P. Myeloid-derived suppressor cells in multiple myeloma: pre-clinical research and translational opportunities. Front. Oncol. 2014;4:348. doi: 10.3389/fonc.2014.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misso G., Zappavigna S., Castellano M., De Rosa G., Di Martino M.T., Tagliaferri P., Tassone P., Caraglia M. Emerging pathways as individualized therapeutic target of multiple myeloma. Expert Opin. Biol. Ther. 2013;13(Suppl 1):S95–S109. doi: 10.1517/14712598.2013.807338. [DOI] [PubMed] [Google Scholar]
- 7.Novak A.J., Darce J.R., Arendt B.K., Harder B., Henderson K., Kindsvogel W., Gross J.A., Greipp P.R., Jelinek D.F. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 8.Claudio J.O., Masih-Khan E., Tang H., Gonçalves J., Voralia M., Li Z.H., Nadeem V., Cukerman E., Francisco-Pabalan O., Liew C.C. A molecular compendium of genes expressed in multiple myeloma. Blood. 2002;100:2175–2186. doi: 10.1182/blood-2002-01-0008. [DOI] [PubMed] [Google Scholar]
- 9.Tai Y.T., Li X.F., Breitkreutz I., Song W., Neri P., Catley L., Podar K., Hideshima T., Chauhan D., Raje N. Role of B-cell-activating factor in adhesion and growth of human multiple myeloma cells in the bone marrow microenvironment. Cancer Res. 2006;66:6675–6682. doi: 10.1158/0008-5472.CAN-06-0190. [DOI] [PubMed] [Google Scholar]
- 10.Rickert R.C., Jellusova J., Miletic A.V. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol. Rev. 2011;244:115–133. doi: 10.1111/j.1600-065X.2011.01067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connor B.P., Raman V.S., Erickson L.D., Cook W.J., Weaver L.K., Ahonen C., Lin L.L., Mantchev G.T., Bram R.J., Noelle R.J. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med. 2004;199:91–98. doi: 10.1084/jem.20031330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez E., Smith E.J., Yashar M.A., Patil S., Li M., Porter A.L., Tanenbaum E.J., Schlossberg R.E., Soof C.M., Hekmati T. The Role of B-Cell Maturation Antigen in the Biology and Management of, and as a Potential Therapeutic Target in, Multiple Myeloma. Target. Oncol. 2018;13:39–47. doi: 10.1007/s11523-017-0538-x. [DOI] [PubMed] [Google Scholar]
- 13.Laurent S.A., Hoffmann F.S., Kuhn P.H., Cheng Q., Chu Y., Schmidt-Supprian M., Hauck S.M., Schuh E., Krumbholz M., Rübsamen H. γ-Secretase directly sheds the survival receptor BCMA from plasma cells. Nat. Commun. 2015;6:7333. doi: 10.1038/ncomms8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oden F., Marino S.F., Brand J., Scheu S., Kriegel C., Olal D., Takvorian A., Westermann J., Yilmaz B., Hinz M. Potent anti-tumor response by targeting B cell maturation antigen (BCMA) in a mouse model of multiple myeloma. Mol. Oncol. 2015;9:1348–1358. doi: 10.1016/j.molonc.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan M.C., Hering M., Peckham D., McDonagh C.F., Brown L., Kim K.M., Meyer D.L., Zabinski R.F., Grewal I.S., Carter P.J. Antibody targeting of B-cell maturation antigen on malignant plasma cells. Mol. Cancer Ther. 2007;6:3009–3018. doi: 10.1158/1535-7163.MCT-07-0464. [DOI] [PubMed] [Google Scholar]
- 16.Tai Y.T., Mayes P.A., Acharya C., Zhong M.Y., Cea M., Cagnetta A., Craigen J., Yates J., Gliddon L., Fieles W. Novel anti-B-cell maturation antigen antibody-drug conjugate (GSK2857916) selectively induces killing of multiple myeloma. Blood. 2014;123:3128–3138. doi: 10.1182/blood-2013-10-535088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hipp S., Tai Y.T., Blanset D., Deegen P., Wahl J., Thomas O., Rattel B., Adam P.J., Anderson K.C., Friedrich M. A novel BCMA/CD3 bispecific T-cell engager for the treatment of multiple myeloma induces selective lysis in vitro and in vivo. Leukemia. 2017;31:2278. doi: 10.1038/leu.2017.219. [DOI] [PubMed] [Google Scholar]
- 18.Seckinger A., Delgado J.A., Moser S., Moreno L., Neuber B., Grab A., Lipp S., Merino J., Prosper F., Emde M. Target Expression, Generation, Preclinical Activity, and Pharmacokinetics of the BCMA-T Cell Bispecific Antibody EM801 for Multiple Myeloma Treatment. Cancer Cell. 2017;31:396–410. doi: 10.1016/j.ccell.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Mikkilineni L., Kochenderfer J.N. Chimeric antigen receptor T-cell therapies for multiple myeloma. Blood. 2017;130:2594–2602. doi: 10.1182/blood-2017-06-793869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali S.A., Shi V., Maric I., Wang M., Stroncek D.F., Rose J.J., Brudno J.N., Stetler-Stevenson M., Feldman S.A., Hansen B.G. T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood. 2016;128:1688–1700. doi: 10.1182/blood-2016-04-711903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotolo A., Caputo V., Karadimitris A. The prospects and promise of chimeric antigen receptor immunotherapy in multiple myeloma. Br. J. Haematol. 2016;173:350–364. doi: 10.1111/bjh.13976. [DOI] [PubMed] [Google Scholar]
- 22.Carpenter R.O., Evbuomwan M.O., Pittaluga S., Rose J.J., Raffeld M., Yang S., Gress R.E., Hakim F.T., Kochenderfer J.N. B-cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma. Clin. Cancer Res. 2013;19:2048–2060. doi: 10.1158/1078-0432.CCR-12-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Agostino M., Boccadoro M., Smith E.L. Novel Immunotherapies for Multiple Myeloma. Curr. Hematol. Malig. Rep. 2017;12:344–357. doi: 10.1007/s11899-017-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shigdar S., Ward A.C., De A., Yang C.J., Wei M., Duan W. Clinical applications of aptamers and nucleic acid therapeutics in haematological malignancies. Br. J. Haematol. 2011;155:3–13. doi: 10.1111/j.1365-2141.2011.08807.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:440. doi: 10.1038/nrd.2017.86. [DOI] [PubMed] [Google Scholar]
- 26.Catuogno S., Esposito C.L., de Franciscis V. Aptamer-Mediated Targeted Delivery of Therapeutics: An Update. Pharmaceuticals (Basel) 2016;9:E69. doi: 10.3390/ph9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catuogno S., Esposito C.L. Aptamer Cell-Based Selection: Overview and Advances. Biomedicines. 2017;5:E49. doi: 10.3390/biomedicines5030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiel W.H., Thiel K.W., Flenker K.S., Bair T., Dupuy A.J., McNamara J.O., 2nd, Miller F.J., Giangrande P.H. Cell-internalization SELEX: method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol. Biol. 2015;1218:187–199. doi: 10.1007/978-1-4939-1538-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catuogno S., Esposito C.L., de Franciscis V. Developing Aptamers by Cell-Based SELEX. Methods Mol. Biol. 2016;1380:33–46. doi: 10.1007/978-1-4939-3197-2_3. [DOI] [PubMed] [Google Scholar]
- 30.Sanda T., Tyner J.W., Gutierrez A., Ngo V.N., Glover J., Chang B.H., Yost A., Ma W., Fleischman A.G., Zhou W. TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 2013;3:564–577. doi: 10.1158/2159-8290.CD-12-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J., Tiemann K., Chomchan P., Alluin J., Swiderski P., Burnett J., Zhang X., Forman S., Chen R., Rossi J. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res. 2013;41:4266–4283. doi: 10.1093/nar/gkt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreaux J., Legouffe E., Jourdan E., Quittet P., Rème T., Lugagne C., Moine P., Rossi J.F., Klein B., Tarte K. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103:3148–3157. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lok C.N., Viazovkina E., Min K.L., Nagy E., Wilds C.J., Damha M.J., Parniak M.A. Potent gene-specific inhibitory properties of mixed-backbone antisense oligonucleotides comprised of 2′-deoxy-2′-fluoro-D-arabinose and 2′-deoxyribose nucleotides. Biochemistry. 2002;41:3457–3467. doi: 10.1021/bi0115075. [DOI] [PubMed] [Google Scholar]
- 34.Catuogno S., Esposito C.L., Condorelli G., de Franciscis V. Nucleic acids delivering nucleic acids. Adv. Drug Deliv. Rev. 2018;134:79–93. doi: 10.1016/j.addr.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Yoon S., Rossi J.J. Emerging cancer-specific therapeutic aptamers. Curr. Opin. Oncol. 2017;29:366–374. doi: 10.1097/CCO.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catuogno S., Rienzo A., Di Vito A., Esposito C.L., de Franciscis V. Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J. Control. Release. 2015;210:147–159. doi: 10.1016/j.jconrel.2015.05.276. [DOI] [PubMed] [Google Scholar]
- 37.Caracciolo D., Montesano M., Altomare E., Scionti F., Di Martino M.T., Tagliaferri P., Tassone P. The potential role of miRNAs in multiple myeloma therapy. Expert Rev. Hematol. 2018;11:793–803. doi: 10.1080/17474086.2018.1517041. [DOI] [PubMed] [Google Scholar]
- 38.Lionetti M., Agnelli L., Lombardi L., Tassone P., Neri A. MicroRNAs in the pathobiology of multiple myeloma. Curr. Cancer Drug Targets. 2012;12:823–837. doi: 10.2174/156800912802429274. [DOI] [PubMed] [Google Scholar]
- 39.Di Martino M.T., Rossi M., Caracciolo D., Gullà A., Tagliaferri P., Tassone P. Mir-221/222 are promising targets for innovative anticancer therapy. Expert Opin. Ther. Targets. 2016;20:1099–1108. doi: 10.1517/14728222.2016.1164693. [DOI] [PubMed] [Google Scholar]
- 40.Zarone M.R., Misso G., Grimaldi A., Zappavigna S., Russo M., Amler E., Di Martino M.T., Amodio N., Tagliaferri P., Tassone P., Caraglia M. Evidence of novel miR-34a-based therapeutic approaches for multiple myeloma treatment. Sci. Rep. 2017;7:17949. doi: 10.1038/s41598-017-18186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nuzzo S., Catuogno S., Capuozzo M., Fiorelli A., Swiderski P., Boccella S., de Nigris F., Esposito C.L. Axl-Targeted Delivery of the Oncosuppressor miR-137 in Non-small-Cell Lung Cancer. Mol. Ther. Nucleic Acids. 2019;17:256–263. doi: 10.1016/j.omtn.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esposito C.L., Nuzzo S., Kumar S.A., Rienzo A., Lawrence C.L., Pallini R., Shaw L., Alder J.E., Ricci-Vitiani L., Catuogno S., de Franciscis V. A combined microRNA-based targeted therapeutic approach to eradicate glioblastoma stem-like cells. J. Control. Release. 2016;238:43–57. doi: 10.1016/j.jconrel.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 43.Abdi J., Jian H., Chang H. Role of micro-RNAs in drug resistance of multiple myeloma. Oncotarget. 2016;7:60723–60735. doi: 10.18632/oncotarget.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y., Li F., Saha M.N., Abdi J., Qiu L., Chang H. miR-137 and miR-197 Induce Apoptosis and Suppress Tumorigenicity by Targeting MCL-1 in Multiple Myeloma. Clin. Cancer Res. 2015;21:2399–2411. doi: 10.1158/1078-0432.CCR-14-1437. [DOI] [PubMed] [Google Scholar]
- 45.Di Martino M.T., Gullà A., Gallo Cantafio M.E., Lionetti M., Leone E., Amodio N., Guzzi P.H., Foresta U., Conforti F., Cannataro M. In Vitro and in Vivo Anti-tumor Activity of miR-221/222 Inhibitors in Multiple Myeloma. Oncotarget. 2013;4:242–255. doi: 10.18632/oncotarget.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morelli E., Biamonte L., Federico C., Amodio N., Di Martino M.T., Gallo Cantafio M.E., Manzoni M., Scionti F., Samur M.K., Gullà A. Therapeutic vulnerability of multiple myeloma to MIR17PTi, a first-in-class inhibitor of pri-miR-17-92. Blood. 2018;132:1050–1063. doi: 10.1182/blood-2018-03-836601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallo Cantafio M.E., Nielsen B.S., Mignogna C., Arbitrio M., Botta C., Frandsen N.M., Rolfo C., Tagliaferri P., Tassone P., Di Martino M.T. Pharmacokinetics and Pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in Mice and Non-human Primates. Mol. Ther. Nucleic Acids. 2016;5:E326. doi: 10.1038/mtna.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pitari M.R., Rossi M., Amodio N., Botta C., Morelli E., Federico C., Gullà A., Caracciolo D., Di Martino M.T., Arbitrio M. Inhibition of miR-21 restores RANKL/OPG ratio in multiple myeloma-derived bone marrow stromal cells and impairs the resorbing activity of mature osteoclasts. Oncotarget. 2015;6:27343–27358. doi: 10.18632/oncotarget.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morelli E., Leone E., Cantafio M.E., Di Martino M.T., Amodio N., Biamonte L., Gullà A., Foresta U., Pitari M.R., Botta C. Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia. 2015;29:2173–2183. doi: 10.1038/leu.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Martino M.T., Amodio N., Tassone P., Tagliaferri P. Functional Analysis of microRNA in Multiple Myeloma. Methods Mol. Biol. 2016;1375:181–194. doi: 10.1007/7651_2015_250. [DOI] [PubMed] [Google Scholar]
- 51.Di Martino M.T., Gullà A., Gallo Cantafio M.E., Altomare E., Amodio N., Leone E., Morelli E., Lio S.G., Caracciolo D., Rossi M. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One. 2014;9:e89659.51. doi: 10.1371/journal.pone.0089659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee L., Bounds D., Paterson J., Herledan G., Sully K., Seestaller-Wehr L.M., Fieles W.E., Tunstead J., McCahon L., Germaschewski F.M. Evaluation of B cell maturation antigen as a target for antibody drug conjugate mediated cytotoxicity in multiple myeloma. Br. J. Haematol. 2016;174:911–922. doi: 10.1111/bjh.14145. [DOI] [PubMed] [Google Scholar]
- 53.Cho S.F., Anderson K.C., Tai Y.T. BCMA CAR T-cell therapy arrives for multiple myeloma: a reality. Ann. Transl. Med. 2018;6(Suppl 2):S93. doi: 10.21037/atm.2018.11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abramson H.N. Monoclonal Antibodies for the Treatment of Multiple Myeloma: An Update. Int. J. Mol. Sci. 2018;19:12. doi: 10.3390/ijms19123924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X.J., Tang Y.M. Cytokine release syndrome in cancer immunotherapy with chimeric antigen receptor engineered T cells. Cancer Lett. 2014;343:172–178. doi: 10.1016/j.canlet.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 56.Foy J.W., Rittenhouse K., Modi M., Patel M. Local tolerance and systemic safety of pegaptanib sodium in the dog and rabbit. J. Ocul. Pharmacol. Ther. 2007;23:452–466. doi: 10.1089/jop.2006.0149. [DOI] [PubMed] [Google Scholar]
- 57.Yu D., Wang D., Zhu F.G., Bhagat L., Dai M., Kandimalla E.R., Agrawal S. Modifications incorporated in CpG motifs of oligodeoxynucleotides lead to antagonist activity of toll-like receptors 7 and 9. J. Med. Chem. 2009;52:5108–5114. doi: 10.1021/jm900730r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.