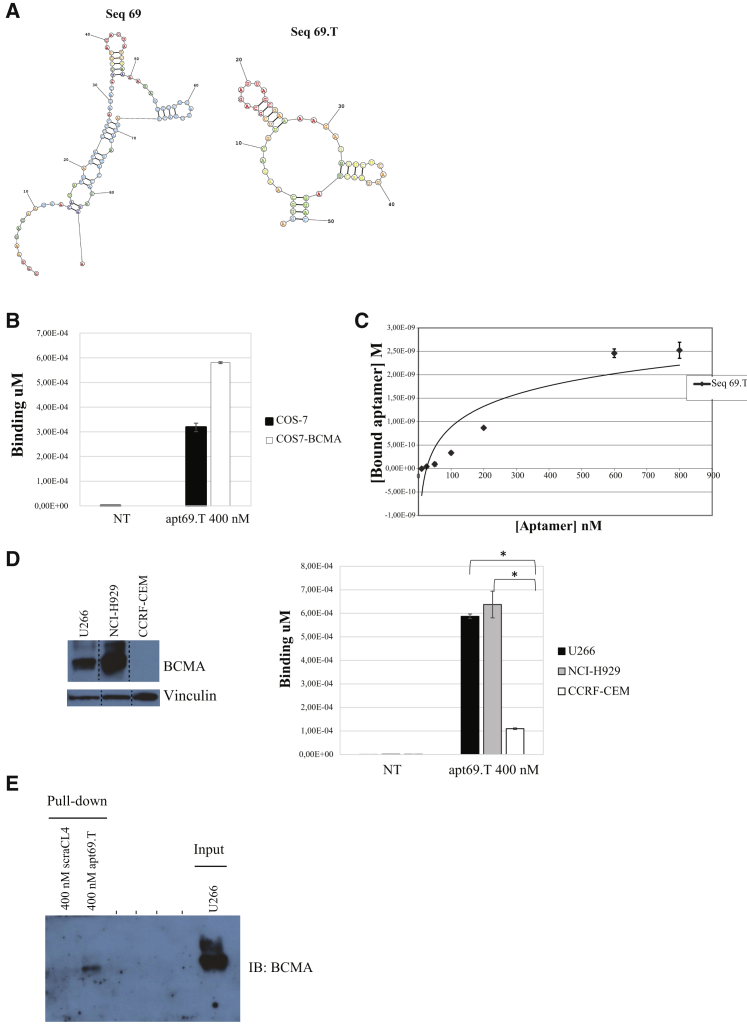
Figure 2.
Apt69.T Specificity
(A) Secondary structure of apt69 full-length aptamer (left) and apt69.T truncated version (right) predicted by RNAstructure Fold. (B) Binding of apt69.T on COS-7 parental cells and COS7-BCMA. Cells were incubated with 200 nM apt69.T for 30 min at 37°C. Binding values (μM) were evaluated by qRT-PCR. Error bars show the mean ± SEM values (C) Determination of the dissociation constant (KD) of apt69.T on COS7-BCMA cells. Parental COS-7 and COS7-BCMA cells were incubated at 37°C with increasing concentrations of the aptamer. The binding data obtained by qRT-PCR were fitted, and KD was calculated. In all binding assays, the background values obtained with an unrelated aptamer were subtracted from the values obtained with the apt69.T. Error bars show the mean ± SEM values. (D) Apt69.T (400 nM) was incubated for 15 min with U266 (BCMA+), NCI-H929 (BCMA+), and CCRF-CEM (BCMA−) cells. Binding values (μM) were evaluated by qRT-PCR. Error bars show the mean ± SEM values. Statistics were calculated using Student’s t test, *p < 0.05. (E) Aptamer-mediated pull-down. U266 cells were incubated with the indicated concentration of biotinylated apt69.T or a control aptamer (scraCL4). Cell lysates were purified on streptavidin beads and immunoblotted with anti-BCMA antibody. 5 μg of total cell extracts (Input) were loaded as a reference.