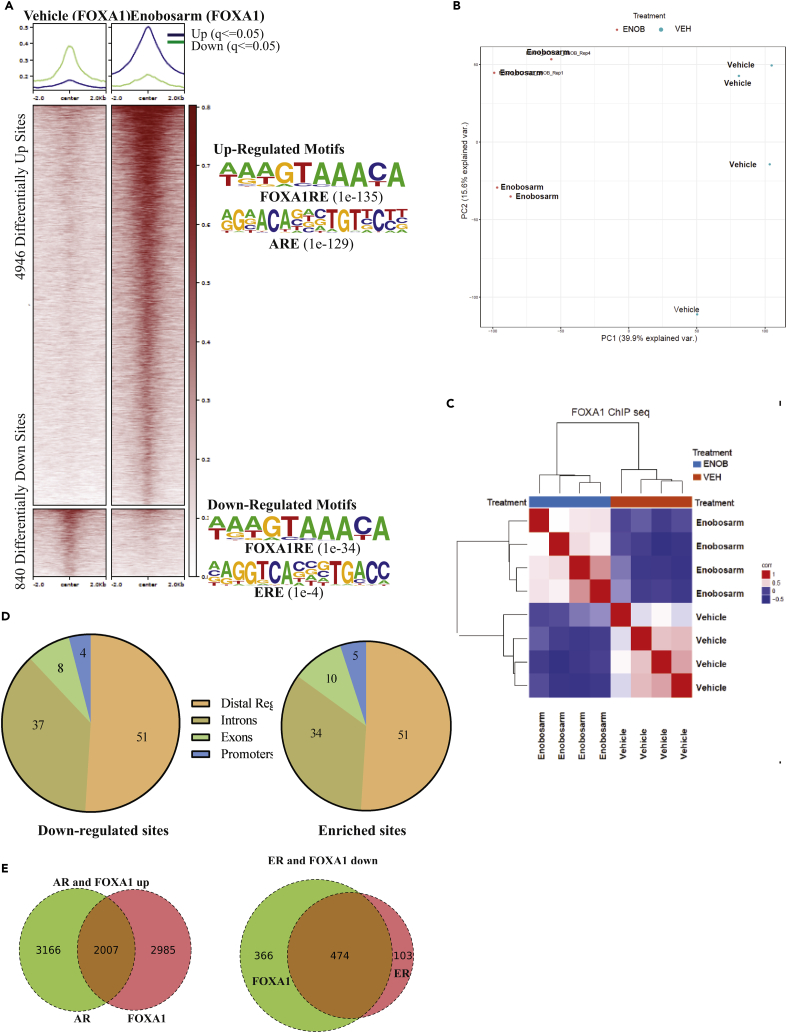
Figure 5.
ChIP-sequencing Shows Reprogramming of FOXA1 Binding to the DNA after Enobosarm Treatment
(A) ChIP assay was performed for FOXA1 in tumors treated with vehicle (n = 4) or 10 mg/kg/day enobosarm (n = 4) (tumors from animals shown in Figure 1D). Next-generation sequencing was performed to determine the genome-wide binding of FOXA1 to the DNA. Heatmap of significantly different peaks (q < 0.05) is shown as average of the individual tumor samples. The top enriched and depleted motifs are shown to the right of the heatmap.
(B) Principal Component Analysis (PCA) plot of vehicle- and enobosarm-treated samples that corresponds to FOXA1-ChIP peaks is shown.
(C) Unsupervised hierarchical clustering.
(D) Pie charts showing the distribution of FOXA1 enrichment in enobosarm-treated HCI-13 samples.
(E) AR and FOXA1 and ER and FOXA1 co-occupied sites represented as Venn diagram. FOXA1, Forkhear box A1; ChIP, chromatin immunoprecipitation; ARE, androgen response elements; GRE, glucocorticoid response elements; FOXA1RE, Forkhead box A1 response element.