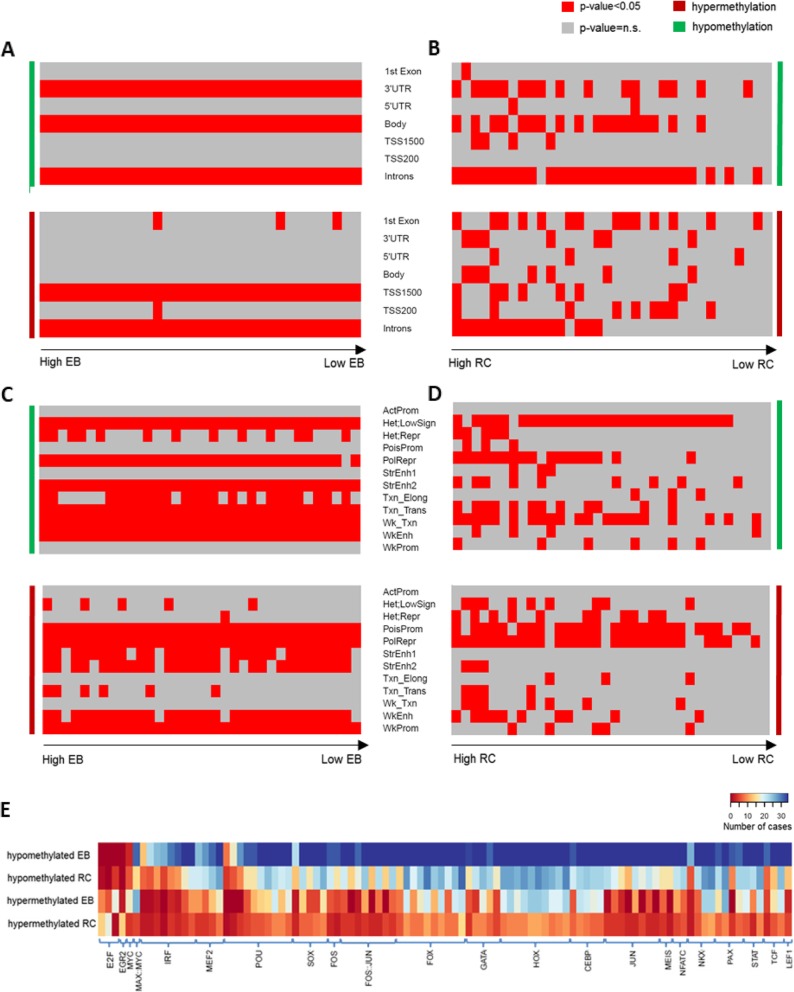
Fig. 2.
Enrichment analysis of the epigenetic burden and relapse changes regarding genomic locations, chromatin states, and transcription factor binding sites. Genomic location enrichment analysis of a the epigenetic burden hypomethylated (green) and hypermethylated (red) CpGs for each CLL case and b the relapse hypomethylated (green) and hypermethylated (red) CpGs for each CLL case. Chromatin states enrichment analysis of c the epigenetic burden hypomethylated (green) and hypermethylated (red) CpGs for each CLL case and d the relapse hypomethylated and hypermethylated (red) CpGs for each CLL case. Each column represents a CLL case, with the cases sorted on x-axis based on the number of DMCpGs per case, from maximum to minimum. Each row represents a genomic element. The red color on heatmap displays the significant enrichment (p < 0.05) in each case for the respective genomic element (ActProm, active promoter; Hete LowSign, heterochromatin low signal; Het Repr, heterochromatin-repressed; PolRepr, polycomb repression; PoisProm, poised promoter; StrEnh1, strong enhancer 1; StrEnh2, strong enhancer 2; Txn_Elong, transcription elongation; Txn_Trans, transcription transition, Wk_Txn, weak transcription; WkEnh, weak enhancer; WkProm, weak promoter. e TFBS analysis of the hypo- and hypermethylated epigenetic burdens and relapse changes revealed significant enrichment for several TFs families (x-axis). The density of heatmap represents the number of patients which showed statistical significant enrichment per TFBS (FDR < 0.05)