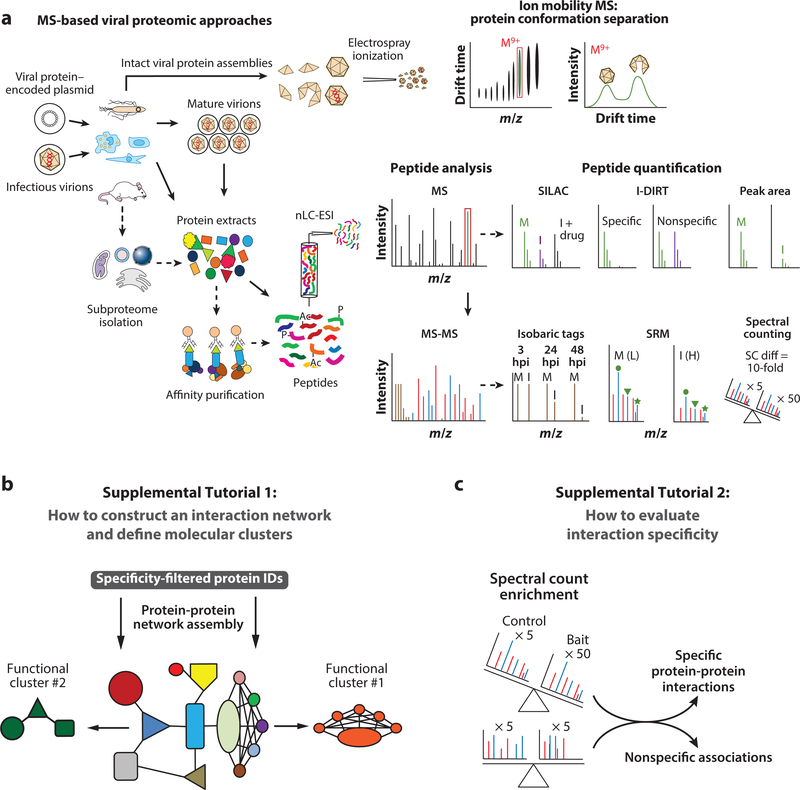
Figure 2.
(a) Multifaceted mass spectrometry (MS)-based analyses of viral structures, interactions, and pathogenesis. Orthogonal workflows illustrate biochemical and MS-based approaches that can be used to study recombinant viral proteins expressed in bacteria or mammalian cells, extracellular virions, or virus-infected mammalian cells. Masses of intact viral assemblies can be determined by electrospray ionization (ESI)–native MS (also called top-down MS), with an orthogonal ion mobility separation to assess molecular compactness (top). Peptide-based MS (also called bottom-up MS) analysis requires denatured protein extracts obtained from extracellular virions, infected cells, organelles, or virus-host complexes. Protein extracts can be directly digested to MS-amenable peptides, or they can first be separated by SDS-PAGE and then in-gel digested into peptides. Following nano–liquid chromatography and ESI (nLC-ESI), peptide sequencing is performed by successive rounds of intact and fragment peptide mass measurements (tandem MS) in the mass spectrometer. Peptide quantification strategies use MS or tandem MS spectra as the basis for protein-level quantification. Global quantitative techniques can use either isotope labeling—within the cell, for simultaneous comparison of usually up to three samples (SILAC), or at the peptide level, for comparison of currently up to ten samples (isobaric tags)—or label-free quantification (peak area or spectral counts). Selected reaction monitoring (SRM) is used for targeted analyses in isotopic or label-free workflows. (b,c) Supplemental Tutorials 1 and 2 provide hands-on experience in the computational analysis of AP-MS data sets. (b) In Supplemental Tutorial 1, a data set of high-confidence protein identifications from HDAC1-EGFP immunoisolations is provided as an example for construction and analysis of functional protein networks using the STRING database of known and predicted protein-protein interactions. (c) In Supplemental Tutorial 2, protein identifications from HDAC1-EGFP immunoisolations that have not been filtered for specificity are provided. The tutorial demonstrates how to use spectral counting to classify specific versus nonspecific associations. Abbreviations: H, heavy; hpi, hours postinfection; I, infected; L, light; M, mock; M9+, molecular ion with a charge state of 9; SC diff, spectral count difference.