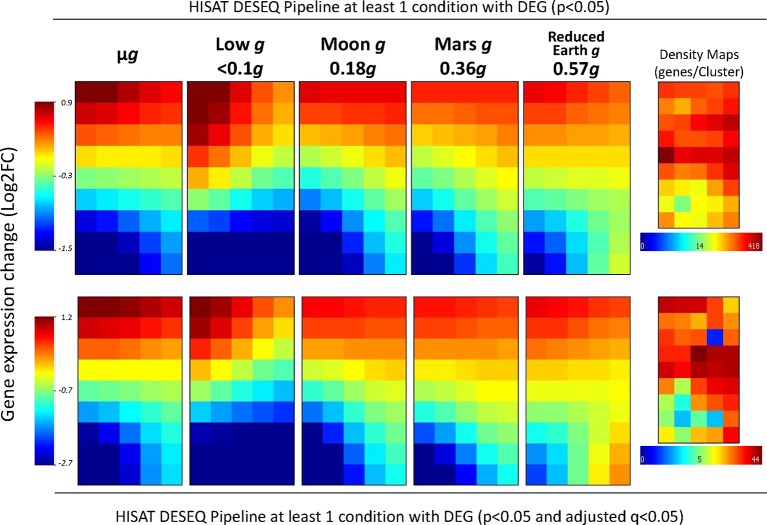
Figure 3.
Whole-genome transcriptional status variations along g-levels into the SG1/SG2 experiments (comparisons versus 1g). A 5x9 clustering analysis of the differentially expressed genes (in at least one of the partial g levels, using normal p values (first row, n = 5,571 genes) or adjusted q-values (second row, n = 861 genes) on the transcriptome have been done with the average of each gene expression level within a similarly expressed cluster across the samples. Values are shown according to the log2 ratio scales at the border of the figure (from highly overrepresented in red to highly down-represented in blue). The mean value of −0.3 or −0.7 indicates an overall repression in gene expression under microgravity. The gene density maps are shown in the middle of the figure for each analysis. Calculated g-levels have been obtained by considering replicates the more similar samples across the nominal µg, 0.1g, 0.3g, 0.5g, 0.8g and 1g (precisely, µg (4 replicates), 0.09 ± 0.02g (3 replicates), Moon level (0.18 ± 0.04g, 3 replicates), Mars-level (0.36 ± 0.02g, 3 replicates), 0.57 ± 0.05g (4 replicates) and control 1g (0.99 ± 0.06g, 3 replicates).