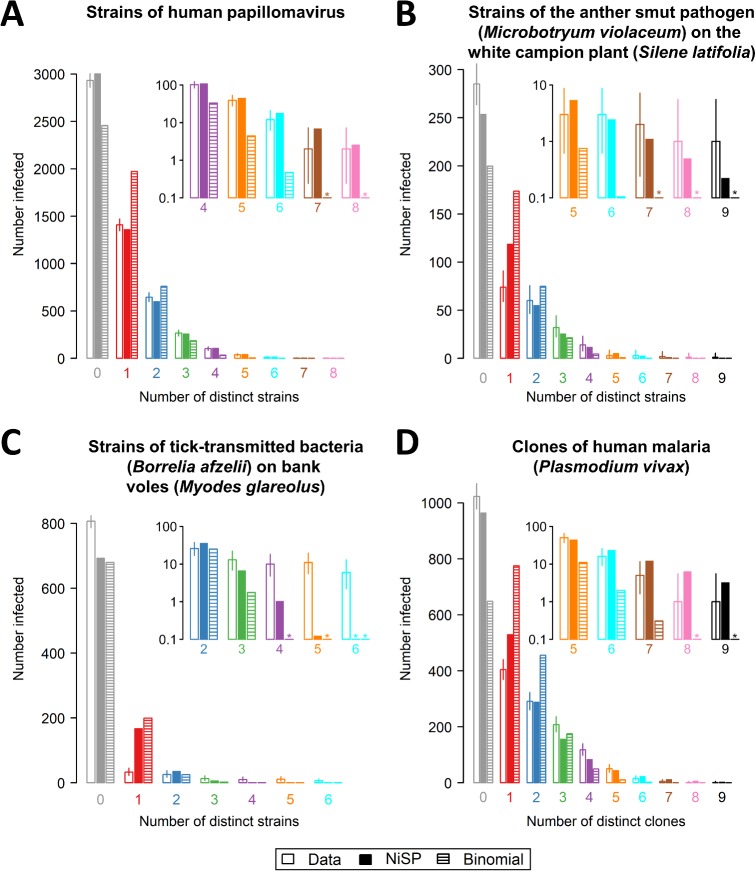
Fig 3. Comparing predictions of the NiSP model with binomial models assuming statistical independence.
In using the NiSP model, pathogens are assumed to be epidemiologically interchangeable: we have therefore restricted attention to data sets concerning strains/clones of a single pathogen species. (A) Strains of human papillomavirus [22]; (B) strains of the anther smut pathogen (Microbotryum violaceum) on the white campion (Silene latifolia) [45]; (C) strains of tick-transmitted bacteria (Borrelia afzelii) on bank voles (Myodes glareolus) [46]; and (D) clones of malaria (Plasmodium vivax) [47]. Insets to each panel show a ‘zoomed-in’ section of the graph corresponding to high multiplicities of clone/strain coinfection, using a logarithmic scale on the y-axis for clarity. Asterisks indicate predicted counts smaller than 0.1. In all four cases, the NiSP model is a better fit to the data than the binomial model (Δ AIC = 572.8,158.6,293.8 and 596.3, respectively). For the data shown in (A), there is no evidence that the NiSP model does not fit the data (lack of goodness of fit p = 0.08), and so our test indicates the human papillomavirus strains do not interact. For the data shown in (B–D), there is evidence of lack of goodness of fit (all have lack of goodness of fit p<0.01). Our test therefore indicates these strains/clones interact (or are epidemiologically different). The underlying data for this figure can be found in S3 Data, S4 Data, S5 Data, and S6 Data. AIC, Akaike information criterion; NiSP, Noninteracting Similar Pathogens.