Figure 4.
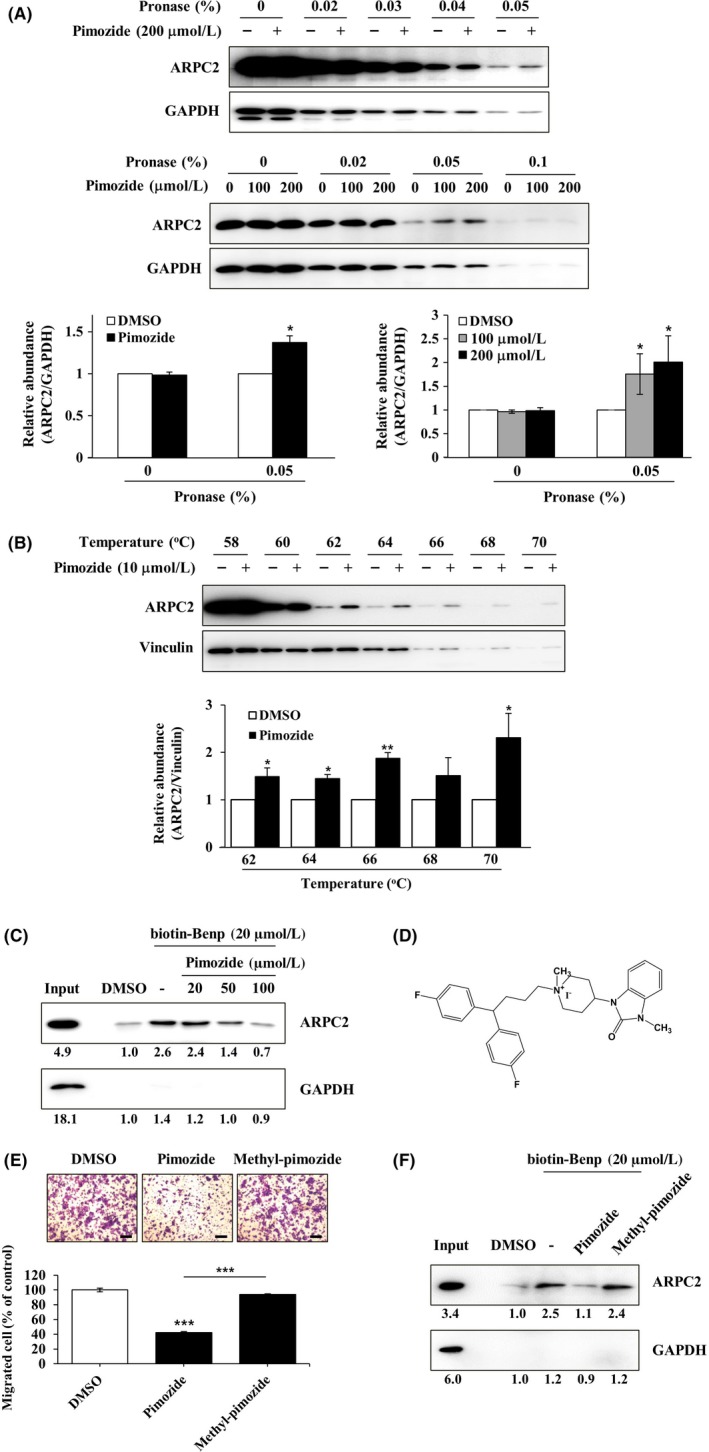
Direct binding of pimozide with ARPC2. A, DLD‐1 cell lysates were incubated in the presence or absence of pimozide (100 or 200 μmol/L) for 1 h at room temperature, followed by proteolysis with various pronases in a dose‐dependent way. GAPDH, which served as the loading control, is relatively resistant to proteolysis (n = 3). B, DLD‐1 cells were treated for 12 h with 10 μmol/L pimozide, and then CETSA was carried out to measure binding ability. Pimozide increased the thermal stability of ARPC2 compared with DMSO. Vinculin is a nontarget protein of pimozide (n = 3). Band intensity was quantified using the MultiGauge program. C, DLD‐1 cell lysates was incubated with 20 μmol/L biotinyl benproperine (biotinyl‐Benp) and competed with pimozide at the indicated concentration. Proteins were captured with NeutrAvidin‐Agarose resin (ThermoFisher Scientific Inc., Waltham, MA, USA) and eluted proteins were analyzed by western blotting (n = 2). D, Chemical structure of N‐methyl‐pimozide. E, Cell migration assay of DLD‐1 cells that were treated with DMSO, pimozide, or N‐methyl‐pimozide for 18 h and quantification of the migrated cells (n = 3). Scale bars, 200 μm. F, Pull‐down assay with biotinyl‐Benp was done in the absence or presence of compounds as competitor (100 μM). Bound proteins on the beads were separated by SDS‐PAGE, and western blot was carried out using anti‐ARPC2 and anti‐GAPDH antibodies (n = 2). Data represent the means ± SD; *P < .05, **P < .01, ***P < .001 compared with the DMSO group
