Figure S1.
RNA Granules Hitchhike on Motile Lysosomes in Mammalian Cells, Related to Figure 1
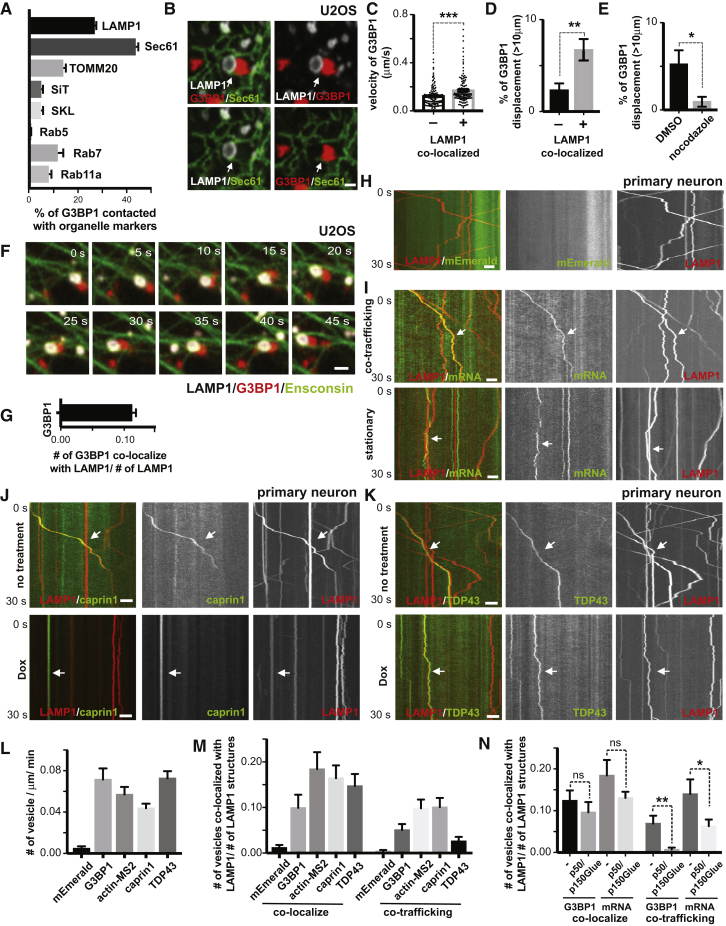
(A) Quantification of the percentage of RNA granules in contact with different organelles from Figure 1A. (n=7).
(B) Contacting RNA granules and lysosomes are frequently in close association with ER. U20S cells were transfected with LAMP1-HaloTag, mEmerald-SEC61 and low levels of mCherry-G3BP1 for 24hrs. Cells were imaged live for 30 minutes after heat shock (43oC). Arrows point to areas where co-localized LAMP1 (white)- and G3BP1 (red)- labeled structures are in close association with Sec61-labeled ER (green). Scale bar: 1μm.
(C) Quantification of velocity of G3BP1 labeled RNA granule co-localized or not co-localized with lysosomes, n= 455 (number of granules, not co-localized), 396 (number of granules, co-localized), t-test, ∗∗∗p < 0.001. Error bars = SEM.
(D) Percentage of G3BP1 labeled RNA granule co-localized or not co-localized with lysosomes with displacement over 10μm, n=7 (number of cells), t-test, ∗∗p < 0.01. Error bars = SEM.
(E) Percentage of G3BP1 labeled RNA granules treated or not treated with nocodazole (Nadezhdina et al. 2010) with displacement over 10μm, n=4, t-test, ∗p < 0.05. Error bars = SEM.
(F) Time-lapse image sequence showing an RNA granule co-trafficking with a lysosome along a microtubule. U2OS cells were transfected with LAMP1-HaloTag, Ensconsin-GFP and low amounts of mCherry-G3BP1 for 24hrs. Images were acquired immediately after heat shock at 43oC. Scale bar: 1μm.
(G) Quantification of LAMP1 labeled lysosomes co-localizing with G3BP1 labeled RNA granules (relative to number of lysosome), n=20 (number of cells).
(H) Kymograph of mEmerald tag and lysosomes in axons. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and PGK promoter driven mEmerald tag. Time-lapse images of axons were acquired at 100ms/frame for 30 seconds. Scale bar: 5 μm.
(I) Kymographs illustrating co-trafficking and stationary interaction patterns of lysosomes with RNA granules. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and actin-24xMBS/MCP-NLS-2xEGFP to label actin mRNA. Upper panel shows co-trafficking of lysosomes and mRNA, and bottom panel shows lysosomes and mRNA associating in a relatively stationary manner. Scale bar: 5 μm.
(J) Kymograph of CAPRIN1-labeled RNA granules co-trafficking with lysosomes in axons.
Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and mEmerald-CAPRIN1 to label RNA granules. Time-lapse images of axons were acquired at 100ms/frame for 30 seconds. Arrows point to lysosomes co-trafficking with CAPRIN1-labeled structures. p50/p150Glued, doxycycline-inducible expression of a p50 dynactin subunit and the CC1 domain of the p150 glued subunit of dynactin. Scale bar: 5 μm.
(K) Kymograph of TDP43-labeled RNA granules co-trafficking with lysosomes in axons.
Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and mEmerald-TDP43 to label RNA granules. Time-lapse images of axons were acquired at 100ms/frame for 30 seconds. Arrow points to a lysosome co-trafficking with a TDP43-labeled structure. Dox, doxycycline-inducible expression of a p50 dynactin subunit and the CC1 domain of the p150 glued subunit of dynactin. Scale bar: 5 μm.
(L) Quantification of frequency of G3BP1, actin-MS2, CAPRIN1, TDP43 labeled RNA granule and mEmerald tag in axons, n=22(mEmerald), 19(G3BP1), 35(actin-MS2), 21(caprin1), 35(TDP43).
(M) Quantification of LAMP1 labeled lysosomes co-localizing or co-trafficking with G3BP1, actin-MS2, CAPRIN1, TDP43 labeled RNA granules and mEmerald tag (relative to number of lysosomes) in axons, n=22(mEmerald), 19(G3BP1), 36(actin-MS2), 25(CAPRIN1), 41(TDP43).
(N) Quantification of LAMP1-labeled lysosomes co-localizing or co-trafficking with G3BP1, actin-MS2 (relative to number of lysosomes), with or without doxycycline-inducible expression of a p50 dynactin subunit and the CC1 domain of the p150 glued subunit of dynactin. N=35(G3BP1,-), 35(G3BP1, p50/p150Glued), 36(actin-MS2, -), 30(actin-MS2, p50/p150Glued). T-test, ∗∗p < 0.01, ∗p < 0.05, ns, not significant. Error bars = SEM.