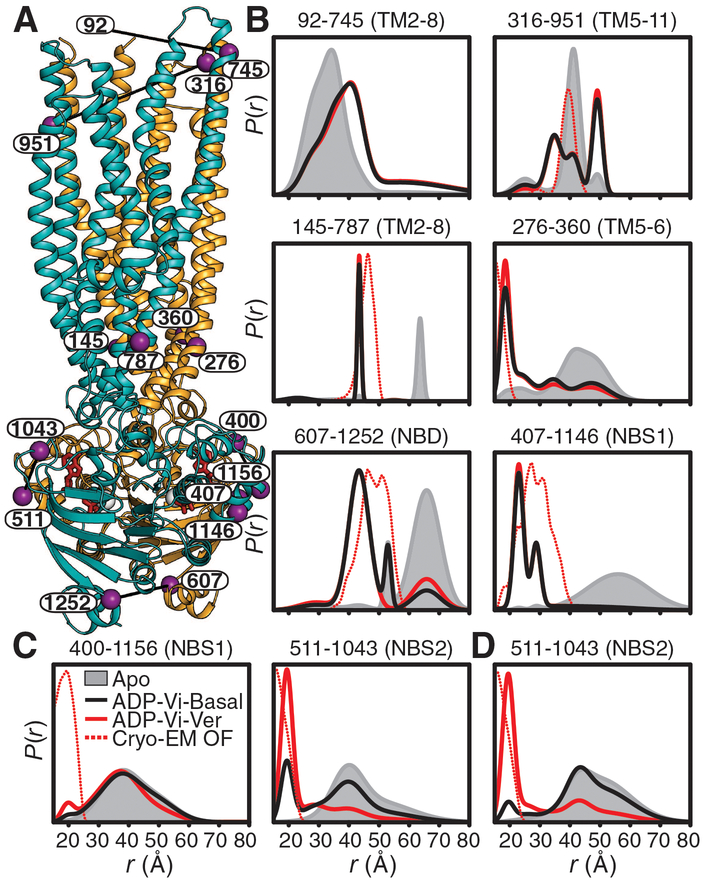
Fig. 1. HESs for basal and substrate-coupled cycles differ by the conformation of the A-loops.
(A) Ribbon representation of the OF Pgp [Protein Data Bank (PDB) code 6C0V] with the N- and C-terminal halves colored with orange and cyan, respectively, and highlighting the positions of spin-label pairs as purple spheres. (B) Distance distributions in the TMDs and NBDs and (C and D) the A-loops obtained in nucleotide-free Pgp (Apo) and the HES (ADP-Vi) in the presence and absence of the substrate Ver in nanodiscs. The corresponding distributions predicted from the cryo-EM OF structure are shown as dashed lines. (D) Distance distributions for NBS2 in mixed micelles are shown for reference. Residue 92 is not resolved in the cryo-EM structure, precluding prediction of distance distributions.