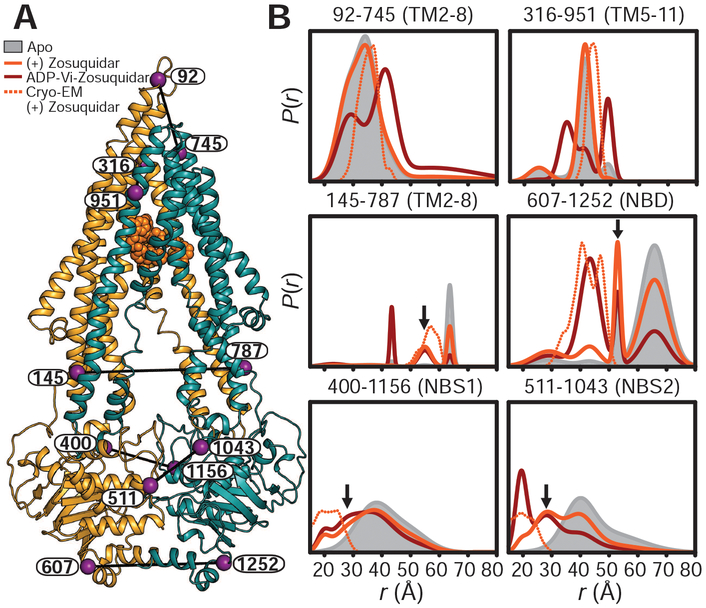
Fig. 3. The high-affinity inhibitor zosuquidar stabilizes a distinct IF conformation of Pgp.
(A) Ribbon representation of zosuquidar-bound Pgp cryo-EM structure (PDB code 6FN1) highlighting the positions of spin-label pairs as purple spheres. (B) Distance distributions obtained in nanodiscs for apo-Pgp, zosuquidar-bound nucleotide-free Pgp, and the HES (ADP-Vi). Predicted distributions are shown as dotted lines. Arrows highlight components on the intracellular side of the TMD and at the A-loops that are either not observed or are minor components in apo-Pgp. The y axes in Figs. 1 and 3 are identical.