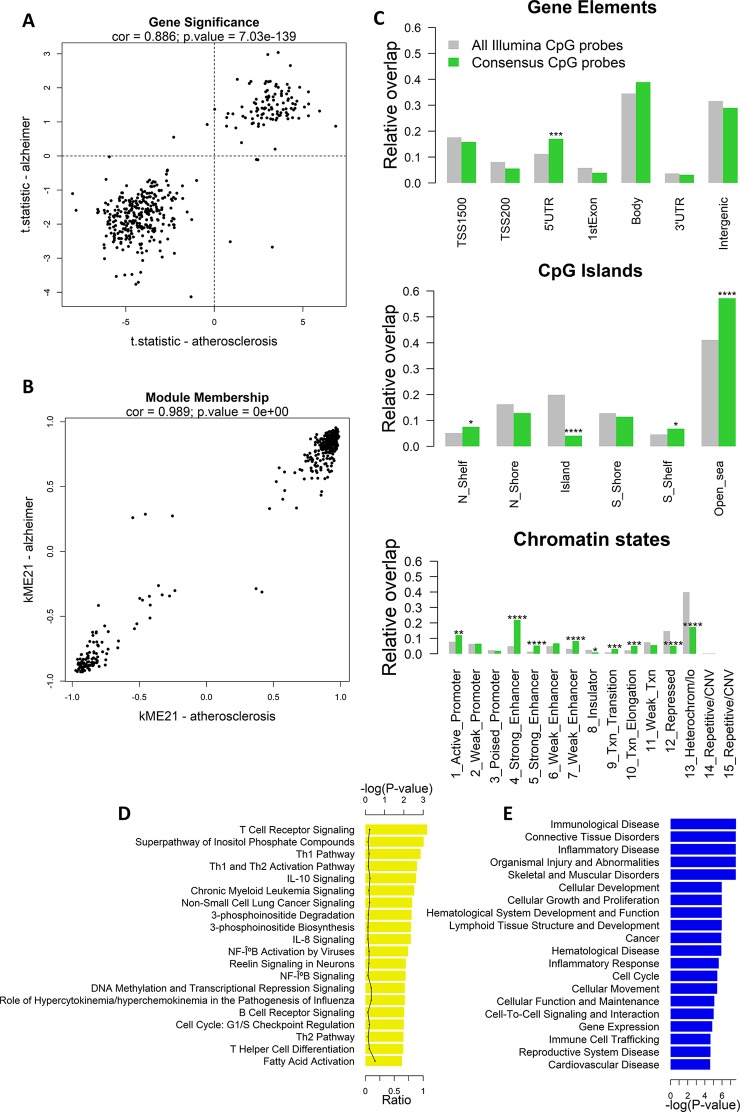
Figure 2.
Weighted Correlation Network Analysis (WGCNA) co-methylation consensus module in atherosclerosis and Alzheimer’s disease (AD) whole blood datasets. (A) Correlation of gene significance values (t-statistics) of CpG probes in the consensus module (module ME21) between atherosclerosis and AD whole blood datasets. The Pearson’s correlation coefficient and p-value are provided at the top of the scatterplot. (B) Correlation of module membership of CpG probes in the consensus module (module ME21) between atherosclerosis and AD whole blood datasets. The Pearson’s correlation coefficient and p-value are provided at the top of the scatterplot. (C) Genomic enrichment of the consensus module CpG probes (module ME21) in multiple genomic regions: gene elements (top), CpG island elements (center), and chromatin segmentation states (bottom). CpG probes in module ME21 were mapped to different genomic regions and enrichment or depletion compared to all Illumina CpG probes was determined using the Fisher’s exact test. Green bars represent the relative overlap of consensus module ME21 CpG probes with the genomic regions, while gray bars represent the relative overlap of all Illumina CpG probes with the genomic regions. * Fisher’s exact P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001. (D) Significantly enriched Ingenuity Pathway Analysis (IPA) canonical pathways, and (E) IPA diseases and biofunctions of genes containing a consensus module CpG probe. Enrichment was calculated using the Fisher’s exact test.