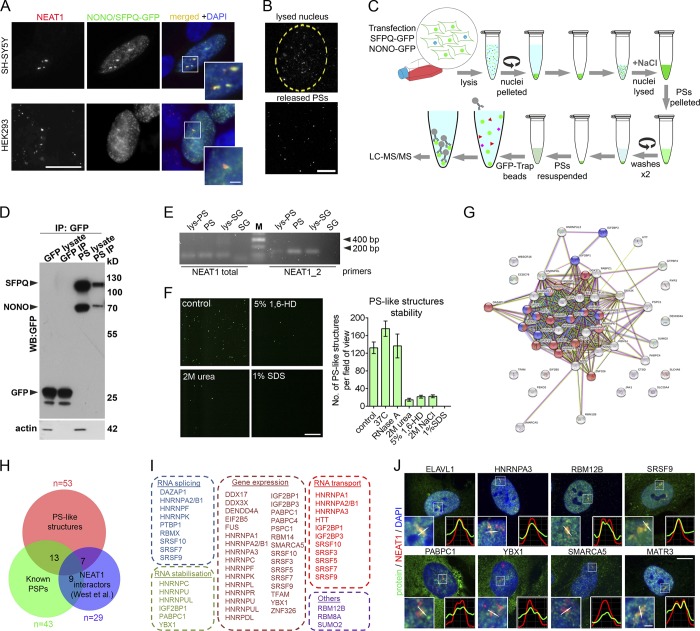
Figure 1.
Purification and proteomic analysis of PS-like structures. (A) PS-like structures formed by coexpressed SFPQ-GFP and NONO-GFP in SH-SY5Y and HEK293 cells contain NEAT1 as revealed by RNA-FISH. Cells were analyzed 24 h after transfection. Scale bars: main image, 10 µm; inset, 2 µm. (B) PS-like structures during the lysis of purified nuclei. Nuclei of SFPQ/NONO-GFP-expressing HEK293 cells were purified and monitored under fluorescent microscope during lysis. Partially lysed nucleus (top, beginning of lysis) and released PS-like structures (bottom, 30 min of lysis) are shown. Scale bar, 20 µm. (C) Workflow used for affinity purification of PS-like structures. (D) Western blot analysis of GFP-Trap bead purified PS-like structures from cells expressing SFPQ/NONO-GFP using an anti-GFP antibody. Samples from cells expressing GFP alone and processed in parallel were used as a control. For lysate, 1/20 of immunoprecipitation (IP) fraction was loaded on gel. (E) Purified PS-like structures but not SG cores contain NEAT1 as revealed by RT-PCR. Lys-PS and lys-SG, total cell lysates of cells used for PS and SG isolation, respectively. (F) Stability of PS-like structures. PS-like structures in the nuclear lysate were treated as indicated for 1 h and imaged under fluorescent microscope (40× magnification). Representative images and quantification of the numbers of GFP-positive dots per field are shown (four to eight fields analyzed). 1,6-HD, 1,6 hexanediol. Error bars represent SEM. Scale bar, 20 µm. (G) STRING functional protein association for the proteome of PS-like structures. In the network, proteins in categories RNA splicing and RNA transport are shown in blue and red, respectively. (H) Overlap between the proteome of PS-like structures, known pool of PSPs (Naganuma et al., 2012; Mannen et al., 2016; Zhang and Carmichael, 2001) and the pool of NEAT1-interacting proteins previously identified by CHART-seq (West et al., 2014). Numbers within circles indicate the number of proteins common between the datasets. Also see Table S1. (I) Biological function enrichment analysis for proteins identified within PS-like structures (also see Table S1, PS-like structures). (J) Validation of proteins identified in the proteomic analysis. Colocalization of proteins and NEAT1 in human fibroblasts was studied using a combination of NEAT1 RNA-FISH and immunocytochemistry. Fibroblasts were treated with MG132 for 4 h to induce PS clusters. MATR3 was included as a positive control. Scale bars: main image, 10 µm; inset, 1 µm.