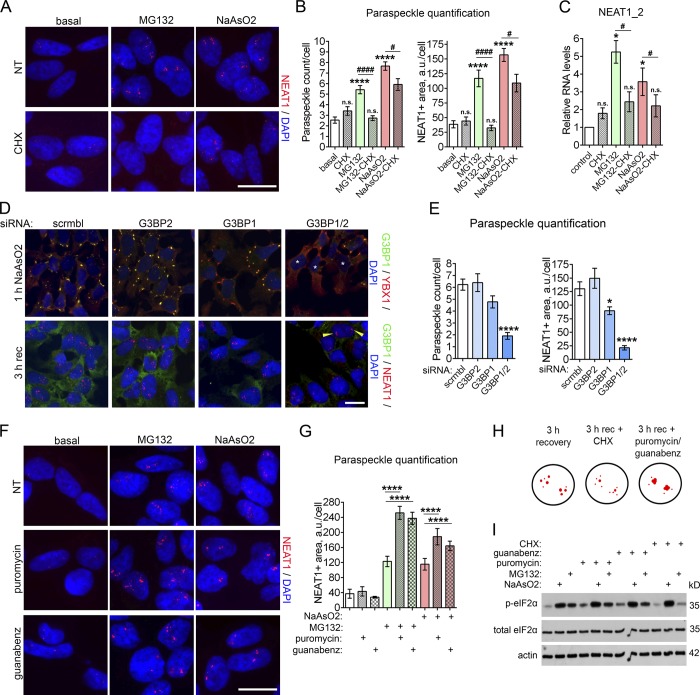
Figure 4.
SGs regulate PS assembly and dynamics. (A–C) Translational inhibitor CHX, which dissipates SGs (also see Fig. S2 A), impedes NEAT1 up-regulation and PS hyperassembly during stress in NaAsO2- and MG132-treated cells. Representative images (A), quantification of PS numbers/area (B), and NEAT1_2 levels (C) are shown. PSs were visualized using NEAT1 RNA-FISH, and NEAT1_2 levels were measured with qRT-PCR. Cells were pretreated with CHX for 1 h before NaAsO2 or MG132 addition to the medium. In B, 107–293 cells were included into analysis. #, P < 0.05; **** and ####, P < 0.0001 (one-way ANOVA with Tukey’s test). In C, n = 3; * and #, P < 0.05 (Mann–Whitney U test). n.s., nonsignificant. (D and E) Depletion of both G3BP proteins disrupts stress-induced PS hyperassembly. Simultaneous siRNA knockdown of G3BP1 and G3BP2 disrupts SG assembly (D, top panel) and reduces NaAsO2-induced PS hyperassembly (D, bottom panel; and E). Representative images (D) and quantification of PS numbers/area (E) are shown. Cells were transfected with respective siRNAs, and 72 h after transfection, treated with NaAsO2. PSs were visualized using NEAT1 RNA-FISH, and SGs using anti-YBX1 staining. Asterisks indicate cells devoid of SGs, and arrowheads indicate intact PS clusters. In E, 178–258 cells were included in the analysis. *, P < 0.05; ****, P < 0.0001 (one-way ANOVA with Tukey’s test). (F and G) Puromycin and guanabenz, which prolong SG presence during stress (also see Fig. S2 D), promote NEAT1 accumulation and impair PS dynamics in stressed cells. Representative images (F) and quantification of NEAT1-positive area (G) are shown. Puromycin or guanabenz was added to the cells simultaneously with MG132 or before recovery from NaAsO2. Cells were analyzed after 4 h of MG132 treatment or after 3 h of recovery from NaAsO2. In G, 67–204 cells were included in analysis per condition. ****, P < 0.0001 (one-way ANOVA with Tukey’s test). (H) Schematic representation of the effect of CHX and puromycin/guanabenz on PSs during recovery from NaAsO2 stress. (I) Levels of p-eIF2α in cells treated with NaAsO2 or MG132 and compounds affecting SG assembly/clearance. Cells were pretreated with CHX for 1 h before NaAsO2 or MG132 addition to the medium. Puromycin or guanabenz was added to the cells simultaneously with MG132 or before recovery from NaAsO2. Cells were analyzed after 4 h of MG132 treatment or after 3 h of recovery from NaAsO2. Note that CHX addition decreases p-eIF2α levels in MG132-treated cells. The experiment was repeated three times, and a representative Western blot is shown. Error bars represent SEM. Scale bars, 10 µm.