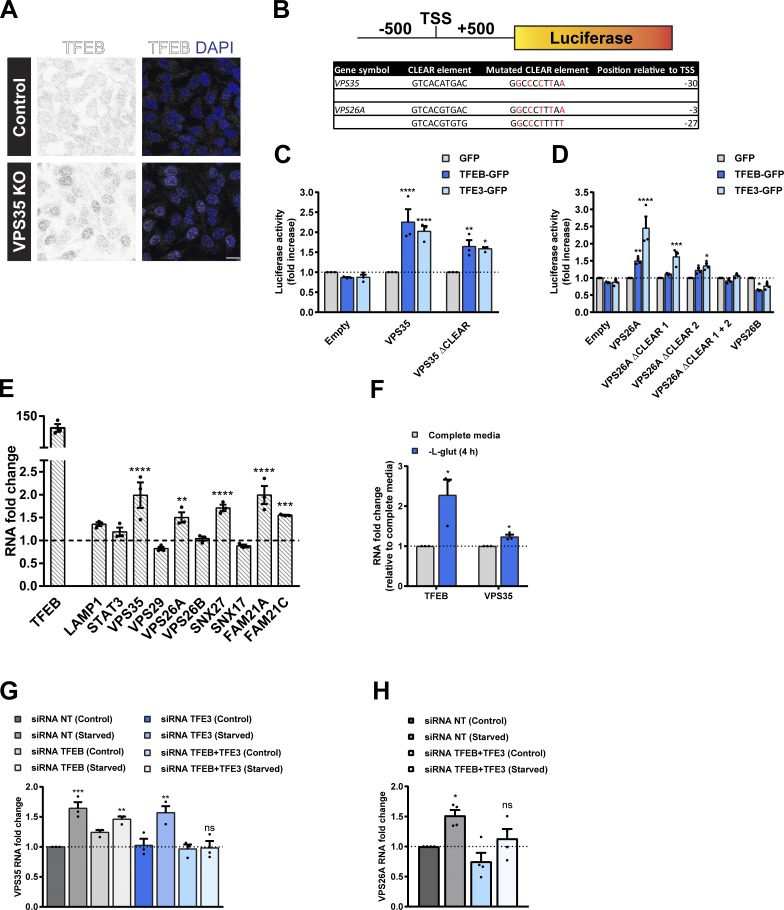
Figure 4.
TFEB and TFE3 promote transcription of the retromer complex in a CLEAR element-dependent manner. (A) Representative IF images of endogenous TFEB in control and VPS35 knockout (KO) HeLa cells. Scale bar, 20 µm. (B) Schematic of the promoter-luciferase reporter constructs used in C and D indicating the location (relative to TSS) and sequence of CLEAR elements in the retromer genes VPS35 and VPS26A and the sequence of the mutated CLEAR elements that were generated. (C and D) Luciferase activity measured in HEK293T cells lentivirally transduced with GFP, TFEB-GFP, or TFE3-GFP and transiently expressing the indicated wild type or mutated CLEAR element VPS35 (C) or VPS26A or VPS26B (D) promoter-luciferase reporter construct. Bars represent a fold increase in luciferase activity relative to cells infected with GFP from three independent experiments; two-way ANOVA followed by Dunnett’s multiple comparison. (E) Quantitative RT-PCR analysis of the retromer complex gene expression in HeLa cells lentivirally transduced with GFP or TFEB-GFP. Data were normalized to ACTB. Bars represent a fold change relative to GFP-transduced cells; n = three independent experiments; two-way ANOVA followed by Sidak’s multiple comparison. (F) mRNA levels of TFEB and VPS35 in HeLa cells treated with L-glutamine starvation for 4 h. Values normalized to ACTB and expressed as a fold change relative to untreated cells (complete media); n = three independent experiments; Student’s t test (unpaired). (G and H) HeLa cells were treated with nontarget siRNA or siRNA to TFEB and/or TFE3 for 72 h. Cells were then maintained in complete media (control) or in EBSS (starved) for 4 h. Total RNAs were extracted, and mRNA transcript abundance was assessed using primers specific for VPS35 (G) or VPS26A (H). Data were normalized to ACTB. Bars represent a fold change relative to nontarget siRNA control cells from at least three experiments; one-way ANOVA followed by Dunnett’s multiple comparison. (C–H) Data represent mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. NT, nontarget.