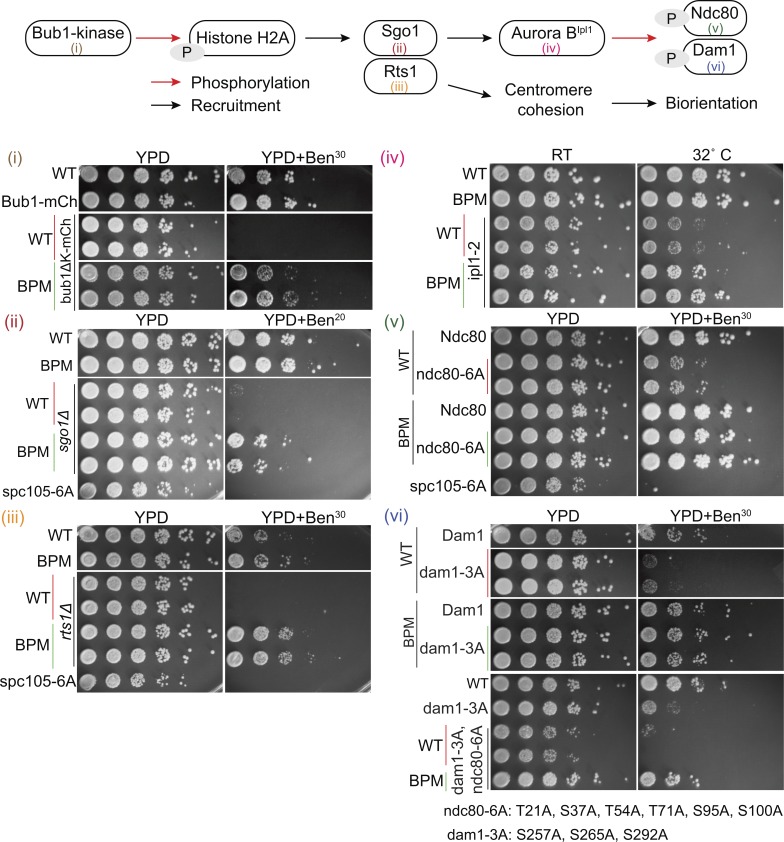
Figure 3.
Spc105BPM suppresses defects in chromosome biorientation and error correction independently of Sgo1. Top: Schematic of the sister centromere cohesion and biorientation pathway in budding yeast. Bottom: Suppression of benomyl sensitivity by Spc105 BPM. Serial dilutions of yeast cells spotted on either rich medium or medium containing either 20 or 30 µg/ml benomyl. WT at the top of each plating indicates WT strain included as a positive control. WT, BPM, or ABPM in other rows refers to the Spc105 allele. spc105-6A, wherein all six MELT motifs are rendered nonphosphorylatable, was used as the negative control.