Abstract
Metastasis-associated macrophages (MAMs) promote persistent growth of breast cancer cells at the metastatic site and are, thus, an attractive therapeutic target to treat breast cancer metastasis, a leading cause of cancer-related death in women. However, the precise mechanisms behind MAM-mediated metastatic tumor outgrowth have not been fully elucidated. Using mouse models of metastatic breast cancer, we showed that MAMs uniquely expressed hepatocyte growth factor (HGF) in metastatic tumors. We also demonstrated that a selected population of cancer cells with high metastatic potential (cancer cells that can establish metastatic tumors in mice with higher number and incidence than parental cells) had higher expression of HGF receptor, MNNG HOS transforming gene (MET), and were more responsive to HGF released from macrophages compared to the parental cells. Blockade of MET signaling in cancer cells suppressed metastatic tumor expansion, in part, through activation of natural killer (NK) cells. Results from this study suggest an approach to prevent life-threatening metastatic tumor formation using blockade of MAM-induced MET signal activation in metastatic cancer cells.
Keywords: breast cancer, hepatocyte growth factor, MET, metastasis, macrophage
Introduction
Breast cancer is a leading cause of cancer-related death in women, largely due to the metastasis in distant organs such as the bone and lung (1,2). Because 6–10% of breast cancer patients present with metastasis at diagnosis (3), novel therapies targeting metastatic tumor outgrowth are necessary (2). Attractive targets are tumor-associated macrophages (TAMs) in the primary and secondary sites (4). However, the roles of TAMs in metastasis are different in the primary and metastatic tumor microenvironment (5). Thus, investigation of TAM functions in metastatic sites is important to develop strategies to block metastatic tumor expansion.
In mouse models of breast cancer pulmonary metastasis, a distinct population of TAMs termed metastasis-associated macrophages (MAMs) promotes extravasation of circulating cancer cells, as well as subsequent survival and persistent growth at the metastatic site (6). However, the mechanisms by which MAMs promote metastatic tumor expansion are not fully understood, and, thus, targetable factors expressed by MAMs need to be identified. Previous animal studies indicate that metastasis promotion by MAMs occurs through the secretion of growth factors. For example, MAMs secrete vascular endothelial growth factor (VEGF) that promotes extravasation of breast cancer cells by increasing vascular permeability, as well as subsequent tumor growth by activating VEGFR1 signaling in MAMs (7,8). It is, therefore, likely that MAM-derived secreted factors play key roles in the metastatic tumor outgrowth. Given that only a minor population of extravasated cancer cells can establish macroscopic metastatic tumors (9,10), we hypothesized that this metastatic population might have been selected to be more responsive to the MAM-derived factors and, thereby, able to expand more efficiently in the metastatic site. Here, we demonstrated that selected populations of mouse mammary and human breast cancer cells with high metastatic potential had higher expression of the hepatocyte growth factor (HGF) receptor, MNNG HOS transforming gene (MET), compared to low metastatic parental cells. This elevated expression resulted in an enhanced and prolonged MET signal activation in response to macrophage-derived HGF. We also showed that the activation of MET signaling in cancer cells promoted metastatic tumor outgrowth via suppression of tumor cell apoptosis induced by natural killer (NK) cells. Our results suggest that blockade of macrophage-mediated MET signal activation could be a potential therapy to prevent metastatic tumor outgrowth of breast cancer.
Materials and Methods
Mice
C57BL/6J, SCID (CB17-Prkdcscid/J), NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ), and human HGF knock-in (HGF-KI) SCID (Hgftm1.1(HGF)AveoPrkdcscid/J) mice were obtained from the Jackson Laboratory. Female SCID mice aged 4 weeks were irradiated with 9 Gy γ-rays and intravenously injected with 4×106 of bone marrow cells from HGF-KI or control SCID mice. The recipient mice were injected with LM2–4175 human breast cancer cells after 6 weeks as described in the ‘Metastasis model’ section. All studies were conducted in accordance with licensed permission from Institutional Animal Care & Use Committee of the Albert Einstein College of Medicine (#20120304) and the UK Home Office (P526C60B3). Sample size to generate significant results (P <0.05) were calculated with on-line software (Openepi).
Healthy donor
Peripheral blood was collected from 5 healthy donors. Blood samples kept on ice were processed to prepare monocyte-derived macrophages within one hour after collection. Informed consent was obtained from all donors included in this study. All study protocols were approved by the IRB of the Albert Einstein Medical College (Bronx, NY, USA).
Cells
E0771 mouse mammary adenocarcinoma cell line was kindly provided by Dr. E. Mihich (Rosewell Park Cancer Institute, NY) in 2010. The LM2–4175 cell line was kindly provided by Dr. J. Massagué (Memorial Sloan Kettering Cancer Center, NY) in 2007. MCF-7 and MDA-MB-231 human breast cancer cells and 3B-11 mouse endothelial cells were obtained from ATCC in 2011. These cells were authenticated and certified by ATCC. All of these cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen) including 10% v/v Fetal Bovine Serum (FBS, Sigma-Aldrich) and 1% v/v Penicillin/Streptomycin (Sigma-Aldrich). We did not culture these cell lines longer than three months after thawing our stocks that were frozen at passage 3 after they were received. Mouse bone marrow-derived macrophages (BMMs) and human monocyte-derived macrophages (MDMs) were isolated as described previously (11,12), and cultured in alpha-Minimal Essential Medium (αMEM, Invitrogen) including 10% v/v FBS and recombinant human CSF1 (1×104 U/mL, Chiron Corporation). Conditioned medium was collected from macrophages cultured in αMEM including 10% v/v FBS and recombinant human CSF1 (1×103 U/mL) for 24 hours. All cells were negative for mycoplasma.
To obtain cancer cell subpopulations with high metastatic potential, we injected 1×106 of E0771 cells into the mammary fat pad or the tail vein of C57BL/6 mice and obtained the lung with metastatic tumors as described in ‘Metastasis models’. The tumor-bearing lungs were minced and incubated with dispase (Gibco) dissolved in Ca2+/Mg2+ free PBS (2.4U/mL) for 30 minutes at 37 °C. The digested tissue was washed by PBS, and cultured in DMEM including 10% v/v FBS and 1% v/v penicillin/streptomycin. The in vivo selection was performed through 2 cycles. The metastasized cancer cells retrieved from spontaneous or experimental metastasis model were named E0771-HML2 or E0771-LG, respectively.
E0771-parental, -HML2, and -LG cells were sub-cloned by limited dilution method. E0771-LG cells were manipulated to express firefly luciferase and transfected with a TRIPZ inducible lentiviral shRNA vector (Dharmacon) including mouse Met shRNA (shMet#1; 5’-GCCAATCTTGCTAAGCAAA-3’ or shMet#2; 5’-GCTACTTATGTGAATGTAA-3’) or non-silencing shRNA (shControl; 5’-CTCGCTTGGGCGAGAGTAA-3’). These cells were cultured in DMEM supplemented with 10% v/v tetracycline-free FBS for their maintenance or with doxycycline (5 μg/mL, Sigma) for shRNA induction up to 4 days.
Metastasis models
In a spontaneous metastasis model, 1×106 of E0771 cells were injected into the mammary fat pad of female C57BL/6J mice (7-weeks-old), and the primary tumor was surgically removed after 4 weeks. Three weeks later, the lung was dissected, and the existence of surface metastatic foci was confirmed under stereoscopic microscopy. The lung with metastatic tumors was used to retrieve cancer cells with higher metastatic potential (E0771-HML2) as described above.
In experimental metastasis models, 1×106 of cancer cells were injected into the tail vein of female mice. E0771-parental, E0771-HML2, and E0771-LG cells expressing shRNA (shCont, shMet#1, and shMet#2) were injected into syngeneic C57BL/6 mice (7-weeks-old). E0771-LG:shMet#1 cells were also injected into SCID and NSG mice (7-weeks-old). LM2–4175 cells were injected into SCID mice that have received bone marrow cells from HGF-KI or control SCID mice as described above. To determine tumor load in the lung area, mice were injected with D-luciferin (GoldBio, 1.5 mg/20 g mouse) into the peritoneum and imaged using Photon Imager Optima (Biospace Lab) every 3–4 days. Photon counts (photon/second/cm2/sr) were quantified by M3 Vision software (Biospace Lab). In some experiments using E0771-LG cells expressing shRNA, we gave doxycycline in the drinking water (SIGMA, 2 mg/mL in 5% w/v sucrose water) or vehicle from 4 days after tumor injection to the experimental endpoint (day 10, 14 or 16 post-tumor injection). In some experiments, we injected blocking antibodies against mouse NK1.1 (PK136; BioXcell, 200 μg/20 g mouse) into the peritoneum on days 4 and 7 after tumor injection.
Spheroid invasion assay
E0771-parental and E0771-HML2 mouse mammary tumor cells or MCF-7, MDA-MB-231, and LM2–4175 human breast cancer cells (5×104) were cultured on top of matrigel (2.5 mg/mL, BD Biosciences) in 35 mm glass bottom dishes, and incubated for 48 hours in αMEM including 10% v/v FBS with or without recombinant mouse/human HGF (10 ng/mL, Peprotech) or undiluted conditioned media (CM) from macrophages (mouse BMMs or human MDMs for mouse and human cancer cells, respectively). In some experiments, cells were incubated with 1 μM crizotinib (SIGMA) or a MET blocking antibody (20 μg/mL; R&D systems). We imaged randomly selected 5 fields with a Zeiss Axioskop II microscope at 10x magnification, and enumerated spheres and invading cells using Fiji software (v1.49, National Institute Health).
In vitro extravasation assay
3B-11 mouse endothelial cells (2×104) were cultured on top of growth factor-reduced matrigel invasion chambers (BD Biosciences) for 48 hours, and BMMs (2×104) were loaded and attached to the bottom of the chambers. The chambers were then placed into a plate with DMEM including 10% v/v FBS and CSF1 (1×104 U/mL). E0771 cells (2×104) labeled with CellTracker CMFDA (Molecular Probe) were loaded into the chambers with DMEM including 0.5% v/v FBS and 1×104 U/mL CSF1. In some experiments, 1 μM crizotinib (SIGMA) was added into the culture. After 36 hours, the chambers were fixed with 4% w/v paraformaldehyde, and cells on top were removed. 5 randomly selected fields in each chamber were imaged by Olympus IX81 fluorescence microscope at 20x magnification, and the number of migrated cancer cells were counted using Fiji software.
Microarray analysis
We extracted RNA from E0771-parental and E0771-HML2 cells using the RNeasy Micro/Mini Kit (Qiagen). RNA samples (15 μg) were hybridized on a GeneChip Mouse Genome 430 2.0 Array and scanned by GeneChip Scanner 3000 (Affymetrix). Datasets were annotated and normalized using the robust multichip average algorithm (rma) from the HuGene 1.0 transcript cluster database (Bioconductor). Genes with FDR ≤ 0.05 and log2 FC ≥ +1.0 were considered differentially expressed. Among genes that were differentially expressed in E0771-HML2, trans-membrane receptor genes were identified as targets by Ingenuity Pathway Analysis software. Data is deposited into GEO (accession number: GSE138139).
Kaplan-Meier plotter database analysis
Using open access software KM Plotter (13), correlation between distant metastasis-free survival (DMFS) of breast cancer patients and expression of target genes (described above) was analyzed. A database was established using multiple microarray data sets downloaded from GEO, and 146 patient data fitting to the selected parameters (Survival: DMFS, Intrinsic subtype: basal, and Grade: 3) were analyzed. For each gene of interest, patients with grade III basal type breast cancer were separated into a high and low expression group based on the median value of the target mRNA expression, and Kaplan-Meier survival curves were plotted. A log-rank P < 0.01 was considered statistically significant. The gene IDs used for the analysis are as follows: 203510_at (MET), 202638_s_at (ICAM1), 215346_at (CD40), 238050_at (ANTXR2), 202803_s_at (ITGB2), 201656_at (ITGA6), 1556582_at (PVR), 203650_at (PROCR), 210845_s_at (PLAUR), 206703_at (CHRNB1), 205718_at (ITGB7), 232317_at (PLXNA4), 218684_at (LRRC8D), 206211_at (ELAM1), 201743_at (CD14), 207072_at (IL18RAP), 204137_at (GPR137B), 209816_at (BCNS), 205891_at (ADORA2B), 204116_at (IL2RG), 219761_at (CLEC1A), and 224392_s_at (OPN3).
Real-time RT-PCR
Total RNA was isolated from untreated cultured cells (BMMs, 3B-11, E0771-parental, -HML2, and -LG cells) using TRI Reagent (Sigma), and cDNA was synthesized from 2 μg of RNA using SuperScript III RT (Life Technologies). Real-time RT-PCR was performed using SYBR master mix (Invitrogen) with the following primers: Gapdh, 5’-AGAACATCATCCCTGCATCC-3’ and 5’-CACATTGGGGGTAGGAACAC-3’; Hgf, 5’-ATGGGGAATGAGAAATGCAG-3’ and 5’-CTCCCTCACATGGTCCTGAT-3’; Met, 5’-ATCATGGGACTTTGCTGGAC-3’ and 5’-ACATTGGGATGGCTG AAGTC-3’. Expression of the target gene was normalized using Gapdh and reported as 2−ΔΔCT.
Western blotting
E0771 cells cultured to 80% confluency were stimulated with recombinant mouse HGF (10 ng/mL, Peprotech) or undiluted BMM-conditioned medium for the indicated time in the presence of 300 nM crizotinib (SIGMA) or vehicle (DMSO, Sigma). Cells were lysed with RIPA Buffer including 1% v/v Halt Protease/Phosphatase Inhibitor (Thermo Scientific), separated by 4–15% w/v SDS gel electrophoresis (Bio-Rad) and transferred onto PVDF membrane (MILLIPORE). The membrane was blocked by Odyssey Blocking Buffer (LI-COR) overnight at 4°C, probed with a primary antibody (1:1000 dilution with blocking buffer) overnight at 4°C, and incubated with horseradish peroxidase-conjugated secondary antibody (1:2000 dilution with blocking buffer) for 1 hour at room temperature after washing with Tris-Buffered Saline Tween-20. Signals were detected using Super Signal Femto (Thermo Scientific) and Odyssey Fc (LI-COR). The following antibodies from Cell Signaling Technology were used: Phospho-Met (#3077P), Met (#4560S), Phospho-Gab1 (#3234T), Gab1 (#3232T), β-actin (#3700S), anti-Rabbit IgG (#7074P2), and anti-mouse IgG (#7076P2).
Flow cytometry
Metastatic lungs were isolated at day 10 after tumor injection from C57BL/6 mice injected with E0771-LG:shMet#1 cells and treated with doxycycline or vehicle as described in ‘Metastasis models’. The lungs were minced by razor blade and incubated with the Lung Dissociation Kit (Miltenyi) at 37 °C for 30 minutes. The single-cell suspensions were washed by PBS including 2% v/w bovine serum albumin (BSA, Sigma), filtered through a 40 μm cell strainer, and treated with RBC lysis buffer (Biolegend). 1×106 of cells were re-suspended in PBS including 2% v/w BSA were blocked with antibody for mouse CD16/CD32 (BD bioscience) on ice for 30 min, and then labeled on ice for 30 min using the following antibodies from Biolegend: anti-CD45 (30-F11), anti-F4/80 (BM8), anti-Ly6G (1A8), anti-CD4 (GK1.5), anti-CD8 (53–6.7), anti-NK1.1 (PK136), anti-CD69 (H1.2F3). Samples from the lung of mice without doxycycline treatment were used as a control.
In order to compare the MET expression, human cancer cells (MCF-7, MDA-MB-231, and LM2–4175) were harvested with 0.05% w/v Trypsin-EDTA (Gibco) and re-suspended in PBS including 2% v/w BSA. 1×106 of cells were labeled with antibodies for human HGFR/c-MET (#95106, R&D) for 30 min on ice. Viability of stained cells was assessed by DAPI stain (Biolegend). The stained cells were detected using LSRII cytometer (BD Biosciences) and analyzed using Flowjo software (TreeStar). The absolute number of cells in tissues was determined as described previously (14).
Histological analysis
To determine the size of metastatic tumors, the lungs were isolated at day 14 or 16 after tumor injection from C57BL/6 mice injected with E0771-LG cells expressing shRNA and treated with doxycycline as described in ‘Metastasis models’. The lungs were fixed with 4% w/v PFA overnight at 4 °C, embedded into paraffin, and sectioned for H&E staining. All lobes were imaged using a Slide Scanner Zeiss Axio Scan Z.1 and analyzed using the Definiens Tissue Studio software to determine average nodule size.
To detect HGF expression in immune cells, the metastatic lungs were isolated at day 10 after tumor injection from C57BL/6 mice injected with E0771-LG cells, and directly embedded in OCT compound (Sakura Finetek). Frozen sections were fixed with cold acetone 5 minutes on ice, washed by cold PBS, and blocked with PBS including 3% v/v BSA and 10% v/v horse serum (Sigma). The sections were then incubated overnight at 4°C with antibodies to mouse HGF (15 μg/mL, R&D) and antibodies (1:100) to Ly6G (1A8), CD3 (17A2), B220 (RA3–6B2) from Biolegend, F4/80 (CI:A3–1, Bio-Rad), or fibroblast antibody (ER-TR7, Novus Biologicals) following 1 hour incubation with fluorescent secondary antibodies (1:500, Alexa555 anti-rat IgG and Alexa488 anti-goat IgG from Molecular Probes) with DAPI (1:250, Invitrogen). Images were taken using Olympus BX61W1 Multiphoton FV1000 Microscope at 20x and 60x magnification. The region with dense DAPI positive staining was outlined as the tumor area, which was confirmed by H&E staining of a serial section.
To detect apoptotic cancer cells, the lungs were isolated at day 10 after tumor injection from C57BL/6 mice injected with E0771-LG:shMet#1 and treated with doxycycline as described in ‘Metastasis models’ and embedded in OCT compound. Sections were fixed and blocked as described above and stained with antibodies (1:100) to ErbB2 (ab16901, abcam) and cleaved caspase 3 (#9664L, Cell Signaling Technology) using the M.O.M. Detection Kit (Vector Labs) and Alexa555 anti-rabbit IgG. Images were taken using Olympus IX73 fluorescence microscope at x10 magnification. The ErbB2+ region (tumor signal) and cleaved caspase 3+ region restricted to the tumor area (apoptosis signal) were quantified using Fiji software (v1.49, National Institute Health). The final apoptotic index was obtained by dividing the apoptosis signal value by the tumor signal value.
MTT assay
E0771-LG cells expressing shRNA (shCont, shMet#1, and shMet#2) were cultured with or without doxycycline (Dox, 5 μg/mL) for 3 days. 1×103 of the pre-treated cells were seeded into 96-well plates with phenol red free DMEM including 10% v/v FBS with or without doxycycline (5 μg/mL). After 3 days, cells were incubated with phenol red free DMEM including 10% v/v FBS and 1.2 mM thiazolyl blue tetrazolium bromide (MTT, Sigma) for 4 hours, and then lysed by equal volume of water with 10 μM hydrochloride and 0.1% w/v SDS (Sigma) to determine absorbance 570 nm.
To investigate effects of MET signal on in vitro cancer cell apoptosis, E0771-LG:shMet#1 cells were cultured with or without doxycycline (5 μg/mL) for 3 days, and 1×103 of these cells were seeded into 96-well plates with phenol red free DMEM including 10% v/v FBS and either recombinant mouse HGF (20 ng/mL), IFNγ (250 U/mL), TNFα (25 ng/mL), TRAIL (200 ng/mL) from Peprotech, or FAS activating antibody (1 μg /mL, JO2, BD Pharmingen). For a low attachment condition, 96-well plates were coated with Poly-2-hydroxethyl methacrylate (poly-HEMA, Sigma). These cells were cultured for 3 days, incubated with MTT, and lysed as described above.
NK cell cytotoxicity assay
NK cells were isolated from the spleen of normal C57BL/6 mice using EasySep Mouse NK Cell Isolation Kit (Stemcell Technologies) and re-suspended in αMEM including 10% v/v FBS. E0771-LG:shMet#1 cancer cells were manipulated to express a red fluorescent protein (mKate) in the nucleus (E0771-LG:shMet#1_NLR) using NucLight Rapid Red Reagent (Essen Bioscience). In some experiments, E0771-LG:shMet#1_NLR were pre-incubated with doxycycline (5 μg/mL; Sigma) or PBS for 3 days and then stimulated with recombinant mouse HGF (20 ng/mL; Peprotech) or PBS for 2 days in the presence or absence of doxycycline (5 μg/mL). In some experiments, conditioned medium was collected from E0771-LG:shMet#1_NLR cells cultured in αMEM including 10% v/v FBS with or without HGF (20 ng/mL) for 2 days. The pre-treated (or untreated) cancer cells were harvested with 0.05% w/v Trypsin-EDTA (Gibco) and resuspended in αMEM including 10% v/v FBS. 1×103 target cancer cells and 4×103 effector NK cells were seeded in 96-well plates coated with basement membrane extract (Geltrex, Gibco), and cocultured in αMEM including 10% v/v FBS (normal medium) or conditioned medium with 1000 U/mL IL-2 (Peprotech) and green fluorogenic caspase-3 substrate (NucView488, Biotium). The cultured cells were imaged by IncuCyte Zoom Live-Cell Analysis System (Essen Bioscience) at x10 magnification for 48 hours, and the number of apoptotic cancer cells (large red/green overlapping nuclei) was counted using the IncuCyte Zoom software (Essen).
Statistical analysis
All samples were collected independently and analyzed by at least two independent experiments. Data were expressed as mean ± SEM and analyzed by Student’s t-test unless it is specified. In some cases, data were analyzed by Chi-square analysis. P <0.05 was considered significant. Statistical analyses were carried out using GraphPad Prism 6.
Results
Cancer cells with high metastatic potential are responsive to macrophage-derived factors
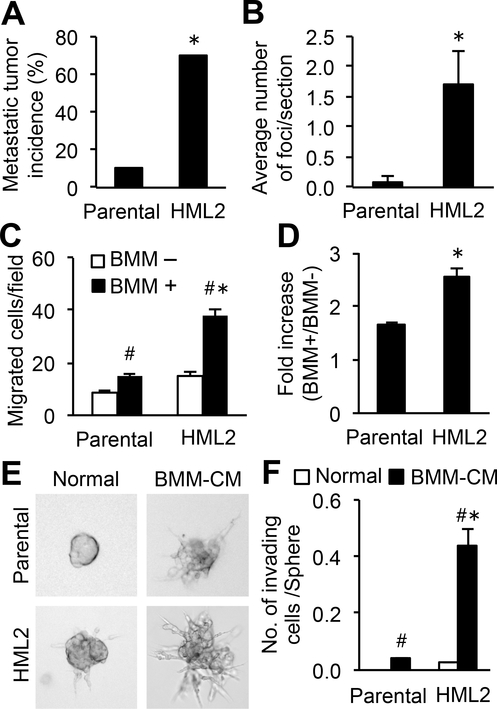
To obtain cancer cell subpopulations with high metastatic potential, we injected E0771 mouse mammary tumor cells (15) into the mammary fat pad of syngeneic C57BL/6 mice and retrieved the metastasized cells from the lung. We repeated this in vivo selection and established a second-generation population (named HML2). This selected population developed pulmonary metastases with a significantly higher incidence and more lesions compared with parental cells (Fig. 1A–B). To compare responses to macrophages between HML2 and parental cells, we performed an in vitro extravasation assay (7,16). The migration of cancer cells through the endothelial monolayer was promoted by bone marrow-derived macrophages (BMMs) placed on the opposite side of the endothelial layer, and this macrophage-promoting extravasation was significantly enhanced in HML2 cells (Fig. 1C–D). We also performed a spheroid invasion assay that recapitulates the invasion of cancer cell clusters into solid tumors (17). Parental E0771 cells formed compact, multicellular tumor spheroids when cultured on high-density matrigel with unconditioned culture medium (DMEM with 10%v/v FBS), whereas the cells cultured in the medium conditioned with BMMs dissociated from the spheroids and invaded into the matrigel (Fig. 1E). The number of invading cells in the presence of BMM-conditioned medium was more than 10-fold higher in HML2 (0.44±0.06) than in parental cells (0.03±0.001) (Fig. 1E–F). These data indicated that the population of E0771 cells with high metastatic potential was more responsive to macrophage-derived secreted factors.
Figure 1. Cancer cell invasion and extravasation induced by macrophage-derived factors are enhanced in a metastatic population of mouse mammary cancer cells.
(A) The incidence of pulmonary metastasis formation in C57BL/6 mice intravenously injected with E0771-parental or their derivative HML2 cells (n=10/group). *P<0.01 versus parental, Chi-square analysis. (B) Average number of metastatic foci in the lungs of mice in A (n=10/group). *P<0.05 versus parental, Student’s t-test. (C) Number of cancer cells transmigrated through an endothelial monolayer in vitro in the presence or absence of bone marrow-derived macrophages (BMMs) and (D) fold increase in the number of transmigrated cancer cells with BMMs relative to that without BMMs (n=5). *P<0.05 versus parental; #P<0.05 versus BMM–, Student’s t-test. (E) Representative morphology of cancer cell spheroids cultured with normal medium or BMM-conditioned medium (BMM-CM) for 48 hours. (F) Average number of invading cells in spheroids (n=5). *P<0.05 versus parental; #P<0.05 versus BMM–, Student’s t-test. Results are mean±SEM.
MET activation by macrophage-derived HGF is enhanced in metastatic cancer cells
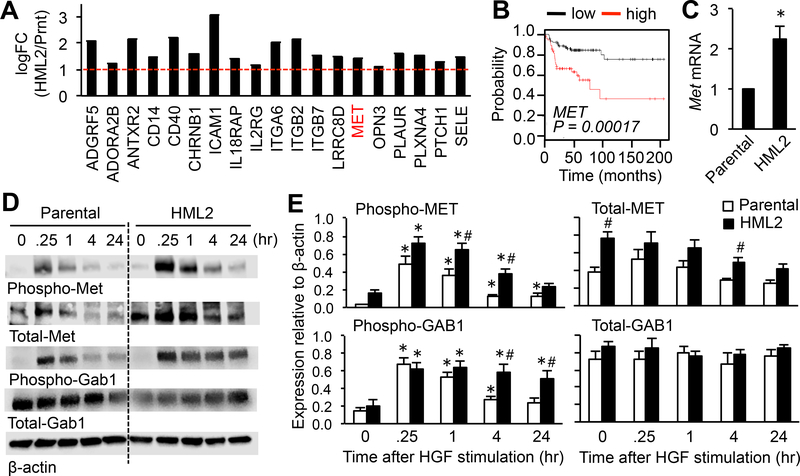
We hypothesized that HML2 compared with parental cells have higher expression of receptors for macrophage-derived ligands that may transmit pro-metastatic signals to the cancer cells. To test this hypothesis, gene expression profiles between HML2 and parental cells were compared by microarray analysis. An unsupervised multi-dimensional scaling plot of all expressed genes showed clear separation of HML2 and parental cells (Supplementary Fig. S1A), and Ingenuity Pathway Analysis identified 19 candidate genes encoding trans-membrane receptors that had high expression (logFC>1.0, P<0.05) in HML2 compared to parental cells (Fig. 2A, Supplementary Fig. S1B). One of these genes encodes MET, an HGF receptor that can promote tumor metastasis in mice (18). Gene set enrichment analysis (GSEA) also suggests that the MET pathway associated genes are differentially expressed by HML2 compared with parental cells (Supplementary Fig. S1C–D). Using a survival analysis tool (13), we further identified that high expression of MET, ATRAB2, and PLAURI, but not others, significantly correlated with lower metastasis-free survival of breast cancer patients (Fig. 2B, Supplementary Fig. S2). Therefore, we hypothesized that enhanced expression of MET can make breast cancer cells more responsive to macrophages and, thereby, promote metastatic tumor expansion.
Figure 2. Expression and activation of MET is increased in the metastatic population of mouse mammary cancer cells.
(A) Genes encoding trans-membrane receptors in microarray data sets that were significantly upregulated (logFC >1, dotted red line, P<0.01) in HML2 compared with parental cells. The scale is exponential. (B) A Kaplan-Meier plot of metastasis-free survival of grade III basal type breast cancer patients with high and low expression of MET. In a database established using multiple microarray data sets downloaded from GEO, 146 patient data fitting to the selected parameters (Survival: DMFS, Intrinsic subtype: basal, and Grade: 3) were analyzed using KM Plotter. (C) Relative Met mRNA expression in parental and HML2 cells (n=3). *P<0.05 versus parental, Student’s t test. (D) Representative images of western blots and (E) total and phosphorylated MET and GAB1 relative to β-actin in parental (Prnt) and HML2 cells stimulated with HGF (n=5) at the indicated time in hours (hr) after HGF stimulation. *P<0.05 versus time 0; #P<0.05 versus parental, Student’s t test. Results are mean±SEM.
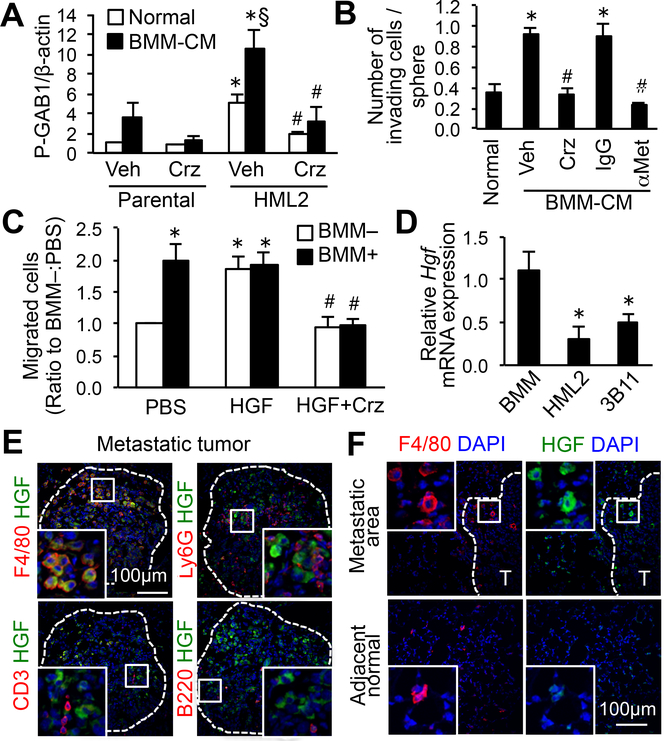
Consistent with the microarray data, we found significantly higher expression of Met mRNA (Fig. 2C) and protein (Fig. 2D–E; total MET) in HML2 cells compared with parental cells. We also found that the ligand-dependent phosphorylation of MET, as well as its downstream target GRB2-associated binder 1 (GAB1), were enhanced and sustained for longer periods in HML2 cells compared with parental cells (Fig. 2D–E). GAB1 activation was induced by conditioned-medium from BMM to a higher extent in HML2 than in parental cells, and this was significantly suppressed by crizotinib, a small molecule MET inhibitor (Fig. 3A). These data indicated that a population of cancer cells with high metastatic potential (i.e., E0771-HML2 cells) expressed high MET that resulted in the enhancement of MET signaling in response to macrophage-derived HGF. We then investigated whether MET activation in HML2 contributed to their hyper-responsiveness to macrophages. In a spheroid invasion assay, invasion of HML2 cells enhanced by BMM-conditioned medium was completely suppressed by crizotinib or an MET neutralizing antibody (Fig. 3B). In an in vitro extravasation assay, BMM-enhanced extravasation of HML2 cells was also suppressed by crizotinib (Fig. 3C). Recombinant HGF alone significantly increased the number of extravasated cancer cells to a comparable level to that induced by BMMs, indicating that HGF was sufficient for cancer cell extravasation (Fig. 3C). These results highlighted the HGF/MET pathway as a pro-metastatic signal that macrophages transmit to metastatic cancer cells. Supporting this conclusion, BMMs expressed higher Hgf mRNA compared with E0771-HML2 cancer cells or 3B11 endothelial cells that were present in the in vitro extravasation assay (Fig. 3D). Using high-resolution immunohistochemical detection, we further identified that HGF expression in E0771 metastatic tumors was detected mainly in F4/80+ macrophages (i.e., MAMs), but not in Ly6G+ neutrophils, CD3+ T cells, or B220+ B cells (Fig. 3E, Supplementary Fig. S3A–B). Consistent with a previous report (19), HGF expression was not found in F4/80+ macrophages in the adjacent normal area that represented resident alveolar macrophages (Fig. 3F). These results showed that MAMs were the major source of HGF in the metastatic tumor microenvironment. Although cancer-associated fibroblasts have been reported to produce HGF (20), we could not find ERTR7+ fibroblasts in the metastatic foci (Supplementary Fig. S3C). Collectively, these results indicated that breast cancer cells with high metastatic potential exhibited high expression of MET and were, thus, more responsive to HGF secreted by MAMs in the metastatic site.
Figure 3. Macrophage-derived HGF activates MET signaling in metastatic cancer cells.
(A) Phospho-GAB1 relative to β-actin in parental and HML2 cells stimulated with normal medium or BMM-conditioned medium (BMM-CM) for 4 hours with MET inhibitor crizotinib (Crz) or vehicle (n=5). *P<0.05 versus parental; §P<0.05 versus normal; #P<0.01 versus vehicle, Student’s t test. (B) Average number of invading cells in E0771-HML2 spheroids cultured with normal medium or BMM-CM for 48 hours in the presence of Crizotinib, vehicle, blocking antibody to MET (αMet), or control IgG (n=9). *P<0.01 versus normal; #P<0.01 versus vehicle/IgG controls, Student’s t test. (C) Number of extravasated E0771-HML2 cells in vitro. Cancer cells were cultured for 36 hours in the presence or absence of BMMs with or without HGF and crizotinib (n=6). *P<0.01 versus PBS:BMM–; #P<0.01 versus HGF, Student’s t test. (D) Relative Hgf mRNA expression in BMMs, E0771-HML2, and 3B11 endothelial cells (n=6). *P<0.01 versus BMMs, Student’s t test. (E,F) Expression of HGF and markers for macrophages (F4/80), neutrophils (Ly6G), T cells (CD3), or B cells (B220) in the lungs of animals with metastatic E0771 tumors (T), and macrophages in normal adjacent tissue (20x magnification). Dotted line indicates a border of metastatic foci (n=3). Scale bar: 100 μm; Square indicates enlarged area at 60x magnification.
Macrophage HGF activates MET in human breast cancer cells to promote metastasis
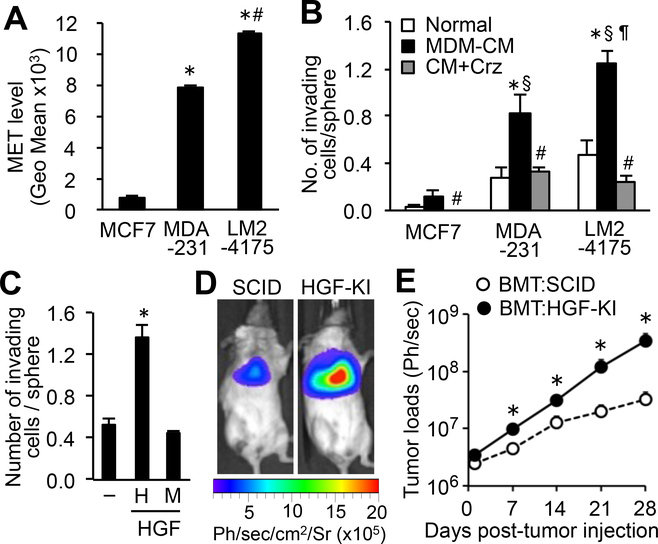
Similar to mouse mammary tumor cells, a highly metastatic subpopulation of MDA-MB-231 human breast cancer cells (LM2–4175)(21) expressed significantly higher MET compared to their parental cells (Fig. 4A). We also found that in vitro invasion of LM2–4175 and MDA-MB-231 was promoted by human monocyte-derived macrophage (MDM)–conditioned medium to a significantly higher extent than in MCF-7 cells that expressed low levels of MET (Fig. 4A–B). The effect of MDM-conditioned medium on tumor cell invasion was completely suppressed by crizotinib (Fig. 4B) and was comparable with that of recombinant human HGF (Fig. 4C). These results indicated that metastatic human breast cancer cells expressed high MET, which enhanced their response to human macrophage-derived HGF.
Figure 4. High MET expression in human breast cancer cells promotes pulmonary metastasis via leukocyte-derived HGF.
(A) Average fluorescence intensity (geometric mean) of MET in the indicated cancer cell lines (n=3). *P<0.01 versus MCF7; #P<0.01 versus MDA-231, Student’s t test. (B) Average number of invading cells in spheroids of the indicated cell lines cultured with normal medium or conditioned medium from human monocyte-derived macrophage (MDM-CM) with or without crizotinib (Crz). Cells were cultured for 48 hours (n=5). *P<0.05 versus normal; §P<0.01 versus MCF7:MDM-CM; ¶P<0.05 versus MDA231:MDM-CM; #P<0.05 versus MDM-CM, Student’s t test. (C) Average number of invading LM2–4175 cells cultured without (–) or with human (H) or mouse (M) HGF for 48 hours (n=5). *P < 0.05 versus HGF–, Student’s t test. (D) Representative images of mice at 28 days post-tumor injection and (E) average tumor load in the lungs (n=4/group). *P<0.05 versus BMT:SCID, Student’s t test. No significant difference at day 0. Results are mean±SEM.
It has been reported that a subpopulation of circulating tumor cells in breast cancer patients express high MET and that patients with high number of MET+ circulating cancer cells show more metastasis and shorter survival than patients with a low number of the cells (22). We, thus, hypothesized that MET activation by MAM-derived HGF was required for circulating breast cancer cells to establish metastatic tumors. To investigate this hypothesis, we injected LM2–4175 breast cancer cells expressing luciferase (21) into the tail vein of immunocompromised SCID mice, a means by which cancer cells are directly introduced into the circulation without being affected by early metastatic steps in the primary site. The recipient SCID mice were irradiated and transplanted with bone marrow cells from human HGF knock-in (HGF-KI) SCID mice in which mouse Hgf coding region is replaced by human HGF cDNA. Thereby, leukocytes in the bone marrow-transplanted (BMT:HGF-KI) mice secrete human HGF, whereas other host cells secrete mouse HGF. Mouse HGF binds to human MET with low affinity (23) and did not signal, as shown by the spheroid invasion assay where LM2–4175 human breast cancer cells did not respond to mouse HGF but showed a robust response to human HGF (Fig. 4C). Therefore, in the metastatic site of BMT:HGF-KI mice, the MET signal in the injected LM2–4175 cells would only be activated by HGF secreted from tumor-infiltrating leukocytes. In support of our hypothesis, the metastatic tumor loads in the lung were significantly higher in the BMT:HGF-KI mice compared with those in control mice without the human HGF gene (Fig. 4D–E). Because only MAMs expressed a significant amount of HGF among tumor-infiltrating leukocytes in mice (Fig. 3E), our results suggest that MAM-derived HGF promotes circulating human breast cancer cells to establish metastatic tumors.
Reduced MET expression suppresses metastatic tumor outgrowth of breast cancer cells
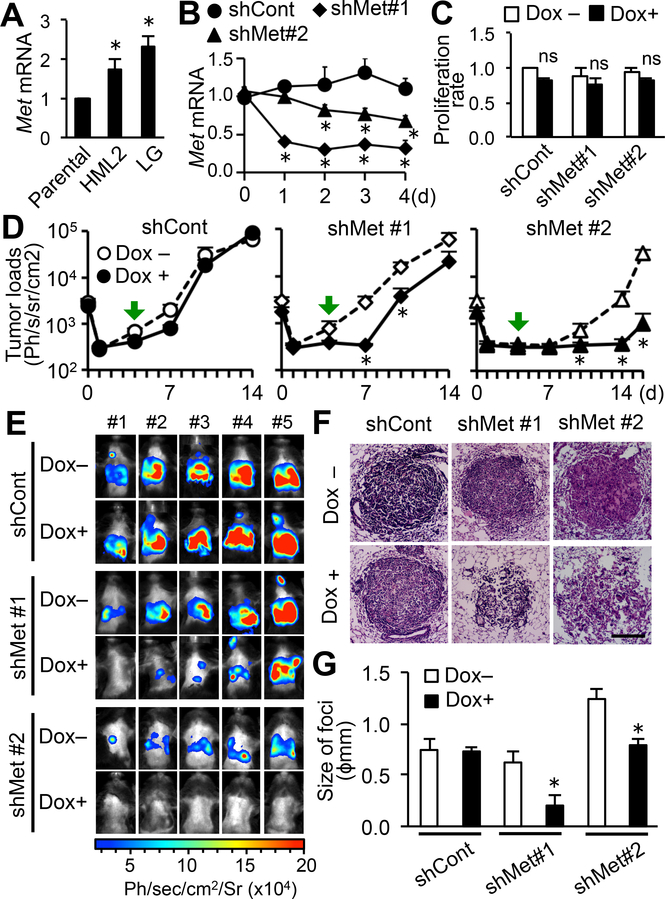
We further investigated whether MET signaling was required for the expansion of circulating tumor cells in the metastatic site. To this end and to indicate our data was not due to clonal variability in the selected HML2 cell line, we used E0771-LG cells, another metastatic derivative of E0771 cells that reproducibly extravasate and establish micro-metastasis via MAM-mediated mechanisms by day 4 and develop into lethal macro-metastasis by day14 after intravenous injection (16,24). Expression of Met mRNA in E0771-LG cells was comparable with that in E0771-HML2 and significantly higher than E0771-parental cells (Fig. 5A). We manipulated E0771-LG cells to express luciferase and an inducible shRNA against Met mRNA (shMet). Expression of Met mRNA in cancer cells with shMet, but not control non-silencing shRNA (shCont), was decreased by doxycycline (Dox) treatment within 2 days without affecting their in vitro proliferation (Fig. 5B–C). We injected the E0771-LG:shMet or E0771-LG:shCont cells into C57BL/6 mice, and treated the animals with Dox from day 4 after tumor injection. As expected, the metastatic loads were not affected by the Dox treatment in mice with E0771-LG:shCont cells. In contrast, the Dox treatment significantly reduced pulmonary tumor burden by day 10 after tumor injection in mice that have received E0771-LG:shMet#1 cells (Fig. 5D). Although tumor load at day 14 in these mice was also lower than that in control mice, it was not statistically significant due to an outlier with significant tumor growth (Fig. 5E). At day 14, we isolated the lung and analyzed H&E sections (Fig. 5F) using a rigorous and accurate stereological method that reads out size of foci (i.e., tumor growth)(6). The Dox treatment also significantly reduced the average size of metastatic tumors established by E0771-LG:shMet#1 cells, whereas it did not affect the size of E0771-LG:shCont tumors (Fig. 5G). We found the reduction in metastatic tumor growth in mice injected with E0771-LG cells expressing another shMet construct (shMet#2), even though metastasis formation of this clone of cells was lower than that of E0771-LG:shMet#1 cells for unknown reasons (Fig. 5D–G). These results showed that breast cancer cells required MET signals for their metastatic tumor outgrowth.
Figure 5. Reduced expression of MET in mouse mammary cancer cells suppresses metastatic tumor expansion in the lung.
(A) Relative Met mRNA expression in E0771-LG and HML2 cells compared with their parental (Prnt) cells (n=6). *P<0.01 versus parental, Student’s t test. (B) Relative Met mRNA expression in E0771-LG cells expressing tetracycline inducible shRNA against Met (shM#1, shM#2) or control non-silencing shRNA (shCont) cultured with doxycycline for 4 days (n=4). *P<0.05 versus day 0, Student’s t test. (C) Cell proliferation measured by MTT assay and expressed as a ratio to doxycycline (Dox) untreated E0771-LG:shCont cells (n=7). ns; not significant. (D) Metastatic tumor load in the lungs of C57BL/6 mice injected with E0771-LG expressing inducible shRNA (shCont, shMet#1, shMet#2) and treated with or without Dox from day 4 after tumor injection (green arrows) to the endpoints (n=5/group). *P<0.05 versus Dox–, Student’s t test. (E) Bioluminescence images of mice shown in D at the experimental endpoint (day 14; shCont and shMet#1 or day 16; shMet#2). (F) Representative H&E stained sections prepared from the lung at day 14 (shCont and shMet#1) or 16 (shMet#2) after tumor injection. Scale bar: 200 μm. (G) Average size of foci in the lungs at day 14 (shCont and shMet#1) or 16 (shMet#2) post-tumor injection (n=5). *P<0.05 versus Dox–, Student’s t test. Results are mean±SEM.
MET prevents metastatic tumor cell apoptosis associated with NK cells
Because metastatic foci established by E0771-LG:shMet cells in Dox-treated mice were sparse in contrast to tightly aggregated foci in control mice (Fig. 5F), we hypothesized that the MET signal might promote metastatic tumor outgrowth by preventing tumor cell apoptosis. We, thus, detected cleaved caspase-3, an apoptosis marker, in the metastatic tumor area indicated by ErbB2 expression (Supplementary Fig. S3D). Although cleaved caspase-3 expression was very low in control tumors, it was significantly increased in tumors established by E0771-LG:shMet cells in Dox-treated mice (Fig. 6A–B), suggesting that disseminated cancer cells require MET signal to suppress apoptosis. To further investigate whether the HGF/Met signal transmits survival signals to cancer cells, we cultured E0771-LG:shMet cells with or without HGF and/or Dox under apoptotic conditions (depletion of serum, prevention of cell attachment, and incubation with recombinant IFNγ, TNFα, TRAIL, and FAS activating antibody). Although these conditions, except for the incubation with TRAIL, significantly reduced viability of cancer cells, neither HGF stimulation nor reduced MET expression altered the extent of tumor cell death induced by these in vitro conditions (Supplementary Fig. S4).
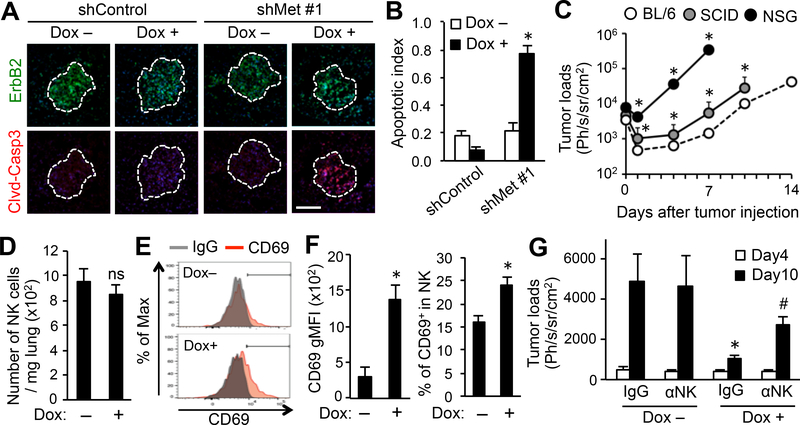
Figure 6. Reduced MET expression increases tumor cell apoptosis partially through a NK cell mediated mechanism.
(A) Expression of markers for cancer cells (ErbB2; green) and apoptosis (cleaved caspase 3; red) in the metastatic lungs of mice at day 10 post-tumor injection. Typical images of tumors at equivalent size are shown. Mice were injected with E0771-LG:shCont or E0771-LG:shMet#1 cells and treated with or without doxycycline (Dox) from day 4 to day 10 post-tumor injection (n=3/group). Dotted line indicates a border of metastatic foci. Scale bar: 200 μm. (B) Apoptotic index of the metastatic foci shown in A (n=3/group; total 30 tumors/group). *P<0.05 versus Dox–, Student’s t test. (C) Metastatic tumor load in the lungs of C57BL/6 (BL/6), SCID, and NSG mice injected with E0771-LG:shMet#1 cells (n=5/group). *P<0.05 versus BL/6, Student’s t test. (D) Number of NK cells in the metastatic lungs of mice at day 10 post-tumor injection. Mice were injected with E0771-LG:shMet#1 cells and treated with or without Dox from day 4 to day 10 post-tumor injection (n=5/group, two independent experiments). ns; not significant. (E) Representative histograms showing CD69 expression in NK cells detected in D. (F) Geometric mean fluorescence intensity (gMFI) of CD69 (left) and percentage of CD69+ cells (right) in NK cells detected in D (n=5/group, two independent experiments). *P < 0.01 versus Dox–, Student’s t test. (G) Metastatic tumor load in the lungs of mice at day 4 and 10 after tumor injection detected by bioluminescence imaging. Mice were treated with or without Dox from day 4 and were injected with a blocking antibody to mouse NK1.1 (αNK) or control IgG at days 4 and 7 after tumor injection (n=5/group). *P<0.05 versus Dox– IgG; #P<0.05 versus Dox+ IgG, Student’s t test. Results are means±SEM.
We, thus, hypothesized that cancer cell apoptosis in the MET inhibited metastatic tumor was mediated by in vivo factors, possibly cytotoxic lymphocytes within the tumors. To investigate this hypothesis, we first evaluated the involvement of CD8+ T and natural killer (NK) cells in metastasis formation. We injected E0771-LG:shMet#1 cells intravenously into SCID mice (lack functional T cells) or NOD/SCID gamma (NSG) mice (lack mature T and NK cells). Although the metastatic tumor growth in the lung was significantly enhanced in SCID mice compared with immune competent C57BL/6 mice, it was further accelerated in NSG mice (Fig. 6C). NK cells induced apoptosis in E0771-LG cells in vitro (Supplementary Fig. S5). Together these results suggest possible roles of NK cells in suppressing metastasis formation. We thus investigated whether reduced MET expression in cancer cells increased accumulation of NK cells within the metastatic lung of C57BL/6 mice, but we did not find a significant difference in the number of NK cells in the lungs between MET expressing (Dox–) and inhibited (Dox+) tumors (Fig. 6D, Supplementary Fig. S6A). We then determined the activation status of these NK cells indicated by CD69 expression (25) and found that CD69 expression and the ratio of CD69+ cells to total tumor-infiltrating NK cells were significantly increased in MET-inhibited tumors compared to MET-expressing tumors (Fig. 6E–F). These results suggest that inhibition of MET expression in cancer cells promotes NK cell activation in the metastatic site.
To further investigate the contribution of NK cells to the growth suppression of MET-inhibited tumors, we injected a NK1.1 blocking antibody or control IgG at days 4 and 7 post-tumor injection into C57BL/6 mice with E0771-LG:shMet#1 tumors with or without Dox treatment from day 4 (Supplementary Fig. S6B). The anti-NK1.1 treatments depleted NK cells in the lung (Supplementary Fig. S6C), and this treatment partially, but significantly, reversed the metastatic tumor load reduced by Dox treatment at day 10 after tumor injection (Fig. 6G), suggesting involvement of NK cells in metastasis suppression caused by MET inhibition. To investigate whether MET activation in cancer cells altered their susceptibility to NK cells, we cultured E0771-LG:shMet cells with NK cells in the presence or absence of HGF and/or Dox, but neither MET knockdown nor HGF stimulation affected the NK cell induced tumor cell apoptosis in vitro (Supplementary Fig. S7A–B). Conditioned medium from E0771-LG:shMet cells cultured with HGF did not suppress NK cell cytotoxicity (Supplementary Fig. S7C–D), suggesting a minor contribution of MET activation in the secretion of NK cell suppressive molecules from cancer cells. Collectively, our data indicated that a population of cancer cells with high metastatic potential upregulate the expression of MET and thereby become more responsive to MAM-derived HGF, and that the enhanced HGF/MET signaling promotes metastatic tumor outgrowth by enhancing trans-endothelial migration and suppression of NK cell functions (Fig. 7).
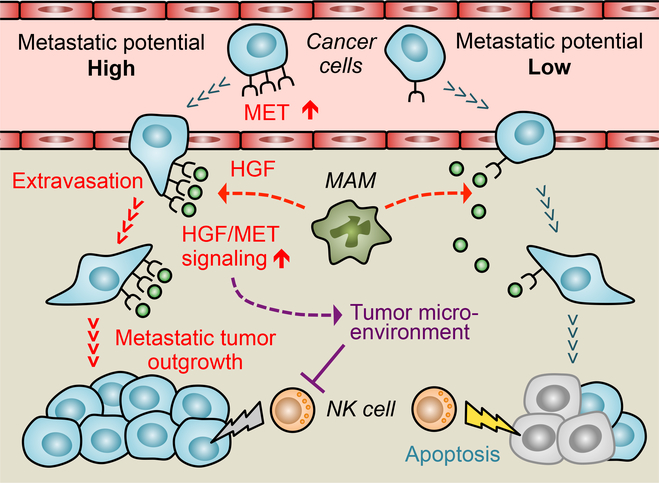
Figure 7. Macrophage activation of MET signaling in cancer cells promotes extravasation and metastatic tumor outgrowth of metastatic breast cancer cells.
Breast cancer cells with high metastatic potential express higher MET, a receptor for hepatocyte growth factor (HGF), than low metastatic cells. This expression results in enhanced and prolonged MET signal activation in response to macrophage-derived HGF. In the metastatic tumor, metastasis-associated macrophages (MAM) express HGF (red dotted arrows), and activation of MET signaling in cancer cells promotes their extravasation, as well as outgrowth, in part, through suppression of natural killer (NK) cell activity (blunted purple line) via affecting the immune suppressive tumor microenvironment (dotted purple line). Gray and yellow lightning bolts represent limited and full NK cell cytotoxicity against cancer cells, respectively.
Discussion
Previous reports show that a subpopulation of circulating tumor cells in breast cancer patients has high MET expression, and this minor population can establish metastatic tumors in immune-deficient mice (22). This data suggests that MET overexpression is advantageous for circulating breast cancer cells to form metastasis. However, very few studies have investigated the requirement of MET signals for cancer cells in the metastatic site. In this study, we identified that mouse mammary and human breast cancer cells, selected for high metastasis rates, expressed high MET and thereby, had enhanced responses to HGF delivered by MAMs. We also showed that MET overexpression in breast cancer cells was required for their trans-endothelial migration in vitro and persistent tumor growth in the lungs of mice. Knockdown of MET in human breast cancer cells also reduces their metastatic expansion in the bone of mice (26). These results indicated that breast cancer cells utilized HGF/MET signal for extravasation and metastatic tumor outgrowth in preferred sites for metastasis.
Although activating mutations and amplification of the MET gene can lead to constitutive MET activation, such genetic alterations are unusual in breast cancer (27). In a mouse model of breast cancer metastasis, blockade of DNA methyltransferase reduces MET expression in cancer cells and suppresses tumor growth in the bone (28). In human breast cancer cells, blockade of DNA methyltransferase increases miR-34–5p expression and concomitantly reduces MET expression (29). Because miR-34-a-5p expression negatively correlates with MET expression in human breast cancer bone metastases (29), it is possible that epigenetic modification of the MET gene induces MET overexpression in metastatic breast cancer cells. Several studies suggest that the MET overexpressing in cancer cells is activated via ligand-dependent mechanisms. For example, MET phosphorylation is scarcely found in a steady state but is increased by HGF stimulation in cultured breast cancer cells originating from patient-derived xenografts (30).
MET and HGF are frequently co-expressed in human breast cancer, and high MET/HGF co-expression associates with high tumor grade (27). Because 59% of MET overexpressing breast cancer expresses HGF in stromal cells (31), it is likely that MET signals in cancer cells is activated in a paracrine manner in a certain number of breast cancers. Several studies have shown that fibroblasts or neutrophils in squamous cell carcinoma or hepatocellular carcinoma secrete HGF and promote invasion of cancer cells (32–34), which indicate these stromal cells as a source of HGF in the tumor microenvironment. However, in our models of breast cancer pulmonary metastasis, MAMs rather than neutrophils expressed high HGF and fibroblasts are rarely found in these metastatic tumors. These results suggest that HGF-producing cells differ between the primary and secondary site and between tumor types. In the current study, we demonstrated that macrophage-derived HGF promoted invasion and extravasation of high metastatic derivatives of breast cancer cells to a higher extent compared with low metastatic parental cancer cells in vitro, and that MET inhibition in cancer cells suppressed metastasis formation in mice. Using the same model, we have shown that metastasizing cancer cells directly interact with MAMs that accumulate in the metastatic site, and that inhibition of the MAM accumulation reduces metastasis formation (6,16). Collectively, our results suggest that the enhanced response to MAMs via an HGF/MET paracrine signal is one of the key characteristics of metastatic breast cancer cells, enabling them to extravasate and subsequently expand in metastatic sites.
We have previously reported that MAMs in the metastatic lung produce VEGF and promote extravasation of metastatic cancer cells through increasing vascular permeability (7). Here, we demonstrated that HGF from macrophages also promoted extravasation of cancer cells. HGF enhances in vitro endothelial adhesion and migration of MDA-MB-231 human breast cancer cells (35,36). However, HGF decreases rather than increases pulmonary endothelial cell permeability in vitro (37). Taken together, MAMs may promote three critical steps in extravasation via secretion of VEGF and HGF: (i) adhesion to endothelial cells, (ii) increasing vascular permeability, and (iii) migration through the vasculature wall (38). In future studies, it will be interesting to determine if MAM-derived HGF contributes to extravasation in different metastatic sites.
MAMs are also reported to promote metastatic tumor outgrowth via suppressing tumor cell apoptosis in the metastatic lung (6). Our results suggest that this process was also promoted via MET signaling. It is reported that MET activation in cancer cells can suppress anoikis and apoptosis induced by nutrient depletion, FAS, and TRAIL (39–42), conditions that circulating tumor cells may encounter during metastasis formation. In our in vitro model, however, HGF did not inhibit tumor cell death under these pro-apoptotic conditions. Instead, our in vivo data suggested that MET activation in cancer cells reduced NK cell-induced apoptosis. A study demonstrates that MET overexpression in renal cell cancer cells increases expression of PD-L1 (43), a ligand of the checkpoint receptor PD-1 that can suppress NK cell cytotoxicity (44). MET signaling prompts lymphoma cells to produce TGFβ (45) that can also suppress NK cell functions (46). These studies suggest that enhanced MET activation in cancer cells can reduce their susceptibility to NK cells via expression of molecules that directly suppress NK cell cytotoxicity. However, HGF stimulation did not reduce apoptosis of E0771-LG mouse breast cancer cells induced by NK cells in vitro, suggesting the involvement of tumor microenvironment mediated by immune suppressor cells such as regulatory T cells and myeloid-derived suppressor cells (47).
Adoptive NK cell transfer has emerged as a novel cancer immunotherapy (48). However, efficacy of this therapy is limited by immunosuppressive tumor microenvironment. It is thus important to identify the mechanisms that modulate NK cell killing capability within the metastatic lesions to improve NK cell-mediated immunotherapy (49). Our results suggest that blockade of MAM-mediated HGF/MET signaling might be an attractive approach to improve NK cell transfer therapy. We have previously reported that the accumulation of MAMs in the metastatic site is promoted by cytokines and chemokines, such as colony-stimulating factor 1 (CSF1), CC-chemokine ligand 2 (CCL2), and CCL3 (5–7,16). Therefore, targeting these cytokine/chemokine receptors in MAMs may reduce the local concentration of HGF in the metastatic tumors. Further investigation regarding synergistic therapeutic effects of antagonists against these cytokine/chemokine receptors in combination with MET inhibitors and NK cell therapy may lead to novel therapies for metastatic breast cancer.
Supplementary Material
Acknowledgments
We thank Dr. Enrico Mihich for providing E0771 cells, Dr. Joan Massagué for providing LM2-4175 cells, Shona Johnston and Will Ramsay for technical support for flow cytometry, and Dr. Sheila Webb for maintenance of mouse colonies. We also thank Dr. Luca Cassetta for critique of the manuscript. This work was supported by grants from National Institutes of Health (RO1#CA172451 and PO1#CA100324 (JWP), USA), the Wellcome Trust (101067/Z/13/Z (JWP), 109657/Z/15/Z (TK), 615KIT/J22738 (TK), UK), and the MRC (MR/N022556/1 (JWP, TK), UK).
Footnotes
Conflict of Interests
The authors declare no competing financial interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Targeting metastasis. Nat Rev Cancer 2016;16:201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawood S, Broglio K, Ensor J, Hortobagyi GN, Giordano SH. Survival differences among women with de novo stage IV and relapsed breast cancer. Ann Oncol 2010;21:2169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitamura T, Qian B-Z, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol 2015;15:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian B, Deng Y, Im JH, Muschel RJ, Zou Y, Li J, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One 2009;4:e6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011;475:222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian BZ, Zhang H, Li J, He T, Yeo EJ, Soong DY, et al. FLT1 signaling in metastasis-associated macrophages activates an inflammatory signature that promotes breast cancer metastasis. J Exp Med 2015;212:1433–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer 2009;9:239–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2:563–72. [DOI] [PubMed] [Google Scholar]
- 11.Tushinski RJ, Oliver IT, Guilbert LJ, Tynan PW, Warner JR, Stanley ER. Survival of mononuclear phagocytes depends on a lineage-specific growth factor that the differentiated cells selectively destroy. Cell 1982;28:71–81. [DOI] [PubMed] [Google Scholar]
- 12.Cassetta L, Kajaste-Rudnitski LA, Coradin T, Saba E, Della Chiara G, Barbagallo M, et al. M1 polarization of human monocyte-derived macrophages restricts pre- and post-integration steps of HIV-1 replication. AIDS 2013;27:1847–56 [DOI] [PubMed] [Google Scholar]
- 13.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725–31. [DOI] [PubMed] [Google Scholar]
- 14.Reeves RK, Evans TI, Gillis J, Wong FE, Connole M, Carville A, et al. Quantification of mucosal mononuclear cells in tissues with a fluorescent bead-based polychromatic flow cytometry assay. J Immunol Methods 2011;367:95–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewens A, Mihich E, Ehrke J. Distant metastasis from subcutaneously grown E0771 medullary breast adenocarcinoma. Anticancer Res 2005;25:3905–16. [PubMed] [Google Scholar]
- 16.Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G, et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med 2015;212:1043–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia 2015;17:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012;12:89–103. [DOI] [PubMed] [Google Scholar]
- 19.Crestani B, Dehoux M, Hayem G, Leçon V, Hochedez F, Marchal J, et al. Differential role of neutrophils and alveolar macrophages in hepatocyte growth factor production in pulmonary fibrosis. Lab Invest 2002;82:1015–22. [DOI] [PubMed] [Google Scholar]
- 20.Comoglio PM, Trusolino L, Boccaccio C. Known and novel roles of the MET oncogene in cancer: a coherent approach to targeted therapy. Nat Rev Cancer 2018;18:341–58. [DOI] [PubMed] [Google Scholar]
- 21.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature 2005;436:518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013;31:539–44. [DOI] [PubMed] [Google Scholar]
- 23.Ikebuchi F, Oka K, Mizuno S, Fukuta K, Hayata D, Ohnishi H, et al. Dissociation of c-Met phosphotyrosine sites in human cells in response to mouse hepatocyte growth factor but not human hepatocyte growth factor: the possible roles of different amino acids in different species. Cell Biochem Funct 2013;31:298–304. [DOI] [PubMed] [Google Scholar]
- 24.Kitamura T, Doughty-Shenton D, Cassetta L, Fragkogianni S, Brownlie D, Kato Y, et al. Monocytes differentiate to immune suppressive precursors of metastasis-associated macrophages in mouse models of metastatic breast cancer. Front Immunol 2018;8:2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fogel LA, Sun MM, Geurs TL, Carayannopoulos LN, French AR. Markers of nonselective and specific NK cell activation. J Immunol 2013;190:6269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Previdi S, Abbadessa G, Daló F, France DS, Broggini M. Breast cancer–derived bone metastasis can be effectively reduced through specific c-MET inhibitor tivantinib (ARQ 197) and shRNA c-MET knockdown. Mol Cancer Ther 2012;11:214–223. [DOI] [PubMed] [Google Scholar]
- 27.Ho-Yen CM, Jones JL, Kermorgant S. The clinical and functional significance of c-Met in breast cancer: a review. Breast Cancer Res 2015;17:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendinelli P, Maroni P, Matteucci E, Desiderio MA. Epigenetic regulation of HGF/Met receptor axis is critical for the outgrowth of bone metastasis from breast carcinoma. Cell Death Dis 2017;8:e2578. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Maroni P, Puglisi R, Mattia G, Carè A, Matteucci E3, Bendinelli P, et al. In bone metastasis miR-34a-5p absence inversely correlates with Met expression, while Met oncogene is unaffected by miR-34a-5p in non-metastatic and metastatic breast carcinomas. Carcinogenesis 2017;38:492–503. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Li S, Wang B, Liu W, Gagea M, Chen H, et al. Cooperative effect of oncogenic MET and PIK3CA in an HGF-dominant environment in breast cancer. Mol Cancer Ther 2019;18:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edakuni G, Sasatomi E, Satoh T, Tokunaga O, Miyazaki K. Expression of the hepatocyte growth factor/c-Met pathway is increased at the cancer front in breast carcinoma. Pathol Int 2001;51:172–8. [DOI] [PubMed] [Google Scholar]
- 32.Knowles LM, Stabile LP, Egloff AM, Rothstein ME, Thomas SM, Gubish CT, et al. HGF and c-Met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clin Cancer Res 2009;15:3740–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, et al. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc Natl Acad Sci U S A 2010;107:11026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He M, Peng A, Huang XZ, Shi DC, Wang JC, Zhao Q et al. Peritumoral stromal neutrophils are essential for c-Met-elicited metastasis in human hepatocellular carcinoma. Oncoimmunology 2016;5:e1219828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mine S, Fujisaki T, Kawahara C, Tabata T, Iida T, Yasuda M, et al. Hepatocyte growth factor enhances adhesion of breast cancer cells to endothelial cells in vitro through up-regulation of CD44. Exp Cell Res 2003;288:189–97. [DOI] [PubMed] [Google Scholar]
- 36.Harrison SM, Knifley T, Chen M, O’Connor KL. LPA, HGF, and EGF utilize distinct combinations of signaling pathways to promote migration and invasion of MDA-MB-231 breast carcinoma cells. BMC Cancer 2013;13:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birukova AA, Cokic I, Moldobaeva N, Birukov KG. Paxillin is involved in the differential regulation of endothelial barrier by HGF and VEGF. Am J Respir Cell Mol Biol 2009;40:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miles FL, Pruitt FL, van Golen KL, Cooper CR. Stepping out of the flow: capillary extravasation in cancer metastasis. Clin Exp Metastasis 2008;25:305–24. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki A, Hayashida M, Kawano H, Sugimoto K, Nakano T, Shiraki K. Hepatocyte growth factor promotes cell survival from fas-mediated cell death in hepatocellular carcinoma cells via Akt activation and Fas-death-inducing signaling complex suppression. Hepatology 2000;32:796–802. [DOI] [PubMed] [Google Scholar]
- 40.Fassetta M, D’Alessandro L, Coltella N, Di Renzo MF, Rasola A. Hepatocyte growth factor installs a survival platform for colorectal cancer cell invasive growth and overcomes p38 MAPK-mediated apoptosis. Cell Signal 2006;18:1967–76. [DOI] [PubMed] [Google Scholar]
- 41.Tang MK, Zhou HY, Yam JW, Wong AS. c-Met overexpression contributes to the acquired apoptotic resistance of nonadherent ovarian cancer cells through a cross talk mediated by phosphatidylinositol 3-kinase and extracellular signal-regulated kinase 1/2. Neoplasia 2010;12:128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du W, Uslar L, Sevala S, Shah K. Targeting c-Met receptor overcomes TRAIL-resistance in brain tumors. PLoS One 2014;9:e95490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balan M, Mier y Teran E, Waaga-Gasser AM, Gasser M, Choueiri TK, Freeman G et al. Novel roles of c-Met in the survival of renal cancer cells through the regulation of HO-1 and PD-L1 expression. J Biol Chem 2015;290:8110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beldi-Ferchiou A, Caillat-Zucman S. Control of NK cell activation by immune checkpoint molecules. Int J Mol Sci 2017;18:E2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumai T, Matsuda Y, Ohkuri T, Oikawa K, Ishibashi K, Aoki N, et al. c-Met is a novel tumor associated antigen for T-cell based immunotherapy against NK/T cell lymphoma. Oncoimmunol 2015;4:e976077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marçais A, Viel S, Grau M, Henry T, Marvel J, Walzer T. Regulation of mouse NK cell development and function by cytokines. Front Immunol 2013;4:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baginska J, Viry E, Paggetti J, Medves S, Berchem G, Moussay E, et al. The critical role of the tumor microenvironment in shaping natural killer cell-mediated anti-tumor immunity. Front Immunol 2013;4:490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol 2016;17:1025–36. [DOI] [PubMed] [Google Scholar]
- 49.Roato I, Vitale M. The uncovered role of immune cells and NK cells in the regulation of bone metastasis. Front Endocrinol 2019;10:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.