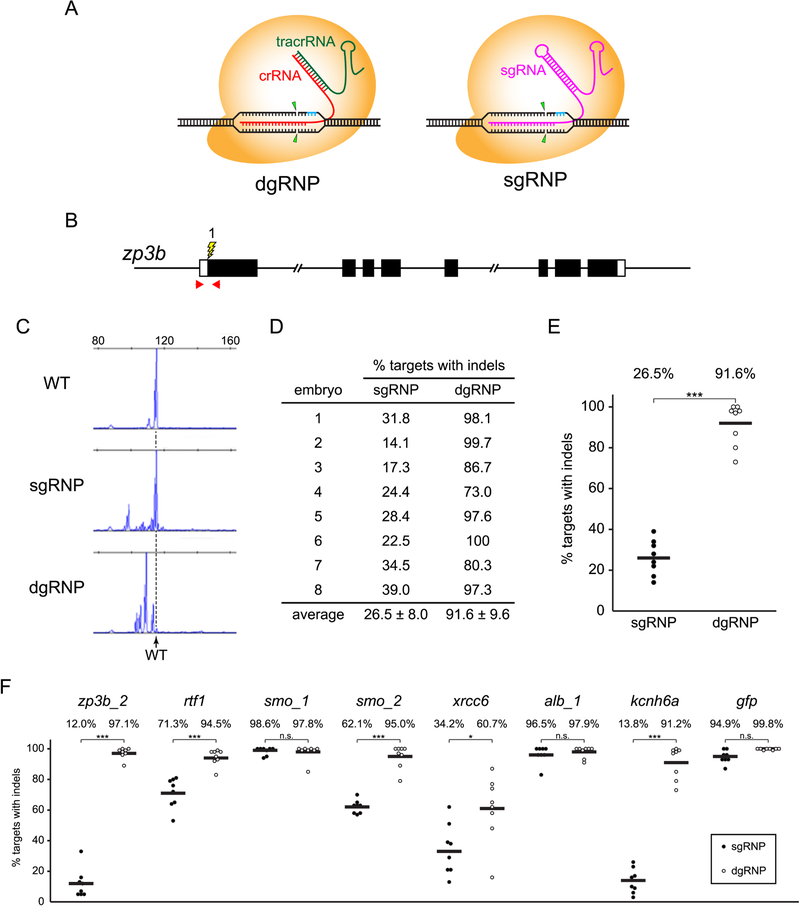
Figure 1.
Experimental approach to compare the mutagenesis activities of sgRNP and dgRNP complexes in zebrafish. (A) Schematic illustration of the CRISPR/Cas9 complexes used in this study. dgRNPs, designed to mimic the native form of the CRISPR/Cas9 complex found in S. pyogenes, consist of the Cas9 protein (orange) with two RNA molecules (crRNA, red, and tracrRNA, green) that form a duplexRNA. The crRNA provides target site specificity. crRNA and tracrRNA molecules were chemically synthesized, often with Alt-R (IDT) modifications to enhance resistance to host cell degradation. sgRNPs consist of the Cas9 protein (orange) with a single gRNA (magenta), designed as a fusion of the crRNA and tracrRNA sequences. Unless otherwise specified, sgRNAs used in this study were generated by in vitro transcription. Both gRNPs require PAM sequences (blue) adjacent to the target site and cleave target sequences similarly (arrowheads), allowing direct comparison of their activities. (B) Target zpb3b_1 (lightning bolt) lies in exon 1, close to the translation initiation site of the gene (boxes indicate exons, filled regions indicate coding sequences). Positions of primer pairs used to amplify the target sequence are indicated (red arrows). (C) Examples of capillary electrophoresis traces of the population of amplicons generated from the genomes of a 24 hpf uninjected WT embryo or embryos that had been injected with gRNPs targeting zpb3b_1. Dashed vertical line indicates mobility of the predominant WT peak. (D, E) Following injection of sgRNPs or dgRNPs into WT eggs, the percent loci with indel mutations was calculated for each embryo (D) and plotted (E). Independent injections yielded highly reproducible results. Statistical differences were calculated using an unpaired Student’s t-test (***p<0.001). (F) dgRNPs are consistently highly mutagenic and their activity exceeds that of sgRNPs at many sites. Fertilized eggs were injected with 1 nl 5μM sgRNP (filled circles) or 5μM dgRNP (open circles) (2.5μM gRNP was injected to target gfp), and the induction of indel mutations was measured by capillary electrophoresis as in (C). Statistical differences were calculated using an unpaired Student’s t-test (*, p<0.05; ***, p<0.001; n.s., not significant). See also Figures S1 and S2.