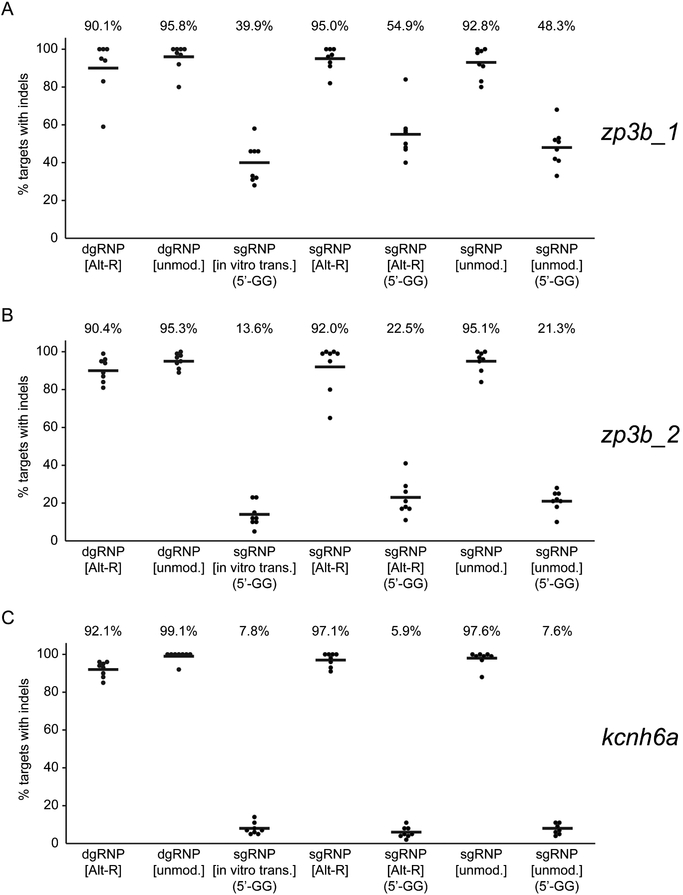
Figure 2.
Supernumerary guanines can reduce sgRNP activity. To identify features of gRNA design and composition that affect gRNP activity, the mutagenic activities of a series of gRNPs targeting (A) zp3b_1, (B) zp3b_2, or (C) kcnh6a were compared. Chemically synthesized duplex guide RNAs were generated with unmodified (unmod.) or Alt-R-modified nucleotides. Single guide RNAs generated by in vitro transcription (in vitro trans.) contained two supernumerary guanine nucleotides (5’-GG) at the 5’ end of the transcript that were not complementary to the target site. Chemically synthesized sgRNAs that perfectly matched the protospacer target sequence or that contained two supernumerary guanine nucleotides (5’-GG) at their 5’ ends were generated with unmodified (unmod.) or Alt-R-modified nucleotides. One-cell embryos were injected with about 1 nl 5μM gRNPs and the induction of indel mutations was measured as in Figure 1. ‘Ideal’ gRNPs composed of gRNAs whose 5’ ends were perfectly complementary to the target site were uniformly highly mutagenic. Each type of gRNP that contained a gRNA with extra 5’ guanine nucleotides was significantly less active than the group of ‘ideal’ gRNPs (p<0.001, One-way ANOVA test).