Abstract
Organic wood extractives—resin acids—significantly contribute to an increase in the toxicity level of pulp and paper industry effluents. Entering open ecosystems, resin acids accumulate and have toxic effects on living organisms, which can lead to the ecological imbalance. Among the most effective methods applied to neutralize these ecotoxicants is enzymatic detoxification using microorganisms. A fundamental interest in the in-depth study of the oxidation mechanisms of resin acids and the search for their key biodegraders is increasing every year. Compounds from this group receive attention because of the need to develop highly effective procedures of resin acid removal from pulp and paper effluents and also the possibility to obtain their derivatives with pronounced pharmacological effects. Over the past fifteen years, this is the first report analyzing the data on distribution, the impacts on living organisms, and the microbial transformation of resin acids. Using the example of dehydroabietic acid—the dominant compound of resin acids in effluents—the review discusses the features of interactions between microorganisms and this pollutant and also highlights the pathways and main products of resin acid bioconversion.
Keywords: diterpenoids, resin acids, biotransformation, biological activity
1. Introduction
The pulp and paper industry is one of the largest industries in the world. According to the UN, pulp production has increased from 163.5 to 183.9 million tons since the beginning of the 21st century [1]. The rise in production capacity is accompanied by an increased pressure on ecosystems, mainly caused by industrial effluent discharge [2].
Produced by coniferous plants of Pinaceae Lindl, resin acids (RAs) are one of the dominant groups of toxic compounds in liquid waste of pulp and paper mills (PPMs) [3]. Because of the shortcomings of the existing treatment systems and high chemical stability of RAs, they accumulate (up to 1500 mg/L) in nearby water bodies. In natural environments, RA concentration decreases due to sorption on solids and bottom sedimentation, as well as due to accumulation in aquatic organisms [4]. Accumulated in tissues and organs of the aquatic biota representatives, RAs cause irreversible negative effects, like red blood cell hemolysis, hepatocellular damage, and ATP depletion of nerve cells [5,6]. RAs can have a strong toxic effect on humans, affecting epithelial cells, polymorphonuclear leukocytes, and gingival fibroblasts [7,8], providing in some cases a pronounced tumor-promoting effect [9] and genotoxicity [10,11].
In this context, it is essential to search for efficient means of ecotoxicant removal. Biotechnological methods of RA conversion are priority solutions in terms of efficiency, safety, and cost-effectiveness. Employing enzymatic activities of microorganisms allows treating these pollutants in one technological stage without the need to use expensive chemicals and under eco-friendly reaction conditions. Despite the obvious toxic effects on living organisms, RAs can be used as parent compounds to derive novel, pharmaceutically significant substances with a broad bioactivity spectrum [12,13].
This review summarizes the data on distribution, impacts on living organisms, neutralization of RAs, and production of compounds with pronounced pharmacological actions. It should be noted that, from the late 1990s [14,15] to the present time, no such work has been carried out. This is the first report analyzing findings on occurrence, effects on aquatic biota, and microbial transformation of RAs over the past fifteen years.
2. Distribution and Toxicity of RAs
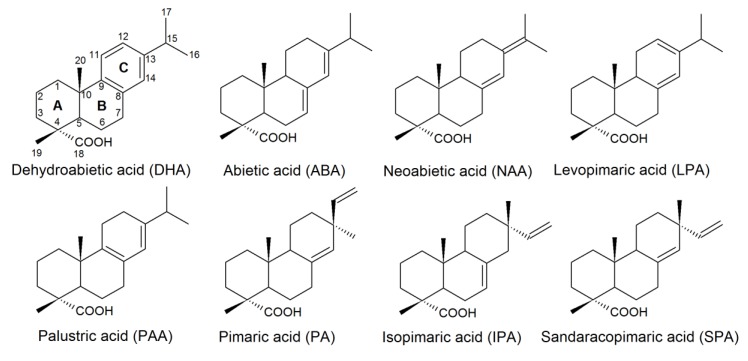
RAs are diterpenic tricyclic monocarboxylic acids represented by two structural stereochemical groups—abietanes and pimaranes. The structures of abietane-type acids include an isopropyl group at the C-13 atom, whereas pimarane-type derivatives have two substituents—vinyl and methyl groups—at the same position [16]. The abietane acids found in PPM effluents contain abietic (ABA), dehydroabietic (DHA), neoabietic (NAA), levopimaric (LPA), palustric (PAA) acids, and pimarane-type compounds include isopimaric (IPA), pimaric (PA), and sandaracopimaric (SPA) acids (Figure 1).
Figure 1.
Structures of resin acids (RAs) found in pulp and paper mill (PPM) effluents.
RAs are major components of lipophilic extractives of generative and vegetative organs of conifers (pine, spruce, fir, and cedar) [17,18,19,20]. The RA content varies depending on the isolation source, season, and climatic conditions for growth of conifers. Thus, RAs and their derivatives are dominant components of fir and spruce seed extractives, being second to only di- and triglycerides [21]. At the same time, ABA dominates (up to 41%) in coniferous plant seeds from Picea, while PAA (up to 35%) is typical for Abies representatives [21].
The quantitative composition of extractives varies according to the object and the seasonal and climatic conditions for growth of coniferous plants [17]. A seasonal dynamics study of RA concentration in the wood of Scots pine (Pinus sylvestris) showed that their concentration increased from 3.17% (July) to 5.39% (January) of the total dry wood weight. The qualitative RA composition is also subject to seasonal variations. During the autumn-winter period, there was an accumulation of DHA from 2.00% (July) to 11.93% (November). A comparative analysis of pine wood samples at various geographical points (from 59° to 68° N) indicated that, towards northward, there was an increase in the total RA content with predominant PA. RAs are supposed to be involved in processes that favor pine adaptation to low temperatures. In addition, antibacterial [22], antifungal [23], and anti-inflammatory [24] properties of RAs protect the trees from pests and various pathogens [25].
The richest (up to 75% of total biomass) source of RAs—the galipot—is released from the damaged bark of conifers. The abundance of RAs in the galipot is variable and also depends on the nature of the source. For example, the Siberian cedar (P. sibirica) galipot is characterized by a relatively uniform content of several (five or more) RAs, while DHA is the dominant component of galipots in most other pine and spruce trees (Table 1) [3,26].
Table 1.
RAs (%) in the galipot of conifers.
| Acid | Scots Pine P. silvestris |
Ordinary Spruce P. excelsa |
Maritime Pine P. pinaster |
|---|---|---|---|
| Abietic | 7.86 | 13.95 | 16.10 |
| Dehydroabietic | 64.58 | 50.08 | 23.50 |
| Pimaric | 10.86 | 7.57 | 10.80 |
| Isopimaric | 8.26 | 18.83 | 6.90 |
| Unidentified | 8.43 | 9.55 | – |
Woodworking and PPMs dealing with coniferous (soft) wood contribute to RA concentration (up to 1500 mg/L) in effluents and their subsequent presence in the environment, posing a toxic effect primarily on the aquatic fauna. The toxicity of most RAs has been studied using test organisms, like daphnia and fish. As seen from Table 2, the acute toxicity values depend on the solubility degree of RAs. In the row DHA ˃ ABA ˃ LPA ˃ NAA ˃ PA ˃ SPA ˃ IPA, solubility decreases from 5.11 to 1.70 mg/L, with IPA being the most toxic to the test organisms [14,27]. It should be noted that pH, temperature, and hardness of receiving waters affect the toxicity and solubility levels of RAs. Zanella [28] reported that when pH changed from 6.5 to 10.0, the acute toxicity level of DHA to daphnia and fish increased up to 76.9 and 45.5 mg/L, respectively.
Table 2.
Solubility and toxicity of RAs.
| RA | Solubility, mg/L | Acute Toxicity (LD50), mg/L | ||||
|---|---|---|---|---|---|---|
| Daphnia Daphnia magna, 48 h |
Rainbow Trout Oncorhynchus mykiss, 96 h |
Red Salmon O. nerka, 96 h |
Silver Salmon O. kisutch, 96 h |
Fathead Minnow Pimephales promelas, 96 h |
||
| DHA | 5.11 | 1.28–6.35 | 0.77–1.32 | 0.50–2.10 | 0.75–1.85 | 2.10–3.20 |
| ABA | 2.75 | 0.68 | 0.72–1.53 | 0.20 | 0.40 | 2.38 |
| LPA | 2.54 | 0.50 | 0.61–1.00 | – | – | – |
| NAA | 2.31 | 0.35 | 0.63–0.71 | – | – | 1.30–1.70 |
| PA | 2.17 | 0.26 | 0.74–1.23 | – | 0.32 | – |
| SPA | 1.82 | 0.13 | – | – | 0.36 | – |
| IPA | 1.70 | 0.07 | 0.40–1.00 | 0.70 | 0.20 | – |
Diluting and discharging into river and sea reservoirs is a widespread approach to PPM effluent disposal [29]. Analysis of filtered PPM effluents showed that only DHA and ABA—as the most soluble RAs—were detected in the water. At the same time, gas chromatography-mass spectrometry of the filtrate registered the presence of RAs, such as DHA, ABA, PA, and NAA, in the sediments [30]. With distance from the industrial effluent discharge sites, the total RA concentration decreased, while DHA concentration increased from 34% to 66% [29]. Thus, DHA is reported as highly resistant to abiotic environmental factors and can be used as an indicator of the open ecosystem pollution by PPM effluents. Undoubtedly, water systems contribute to reduced RA concentrations due to dilution. However, some authors believe that RAs (because of their lipophilic nature) are rapidly adsorbed on suspended solids and characterized by an increased settling capacity [29].
A decrease in RA concentration in water bodies is possible due to bioaccumulation of the latter in hydrobionts [4]. Concentration of RAs in fish can exceed the same value in water [31]. RAs were found in various organs and tissues of marine and river fish. The most significant accumulation was recorded in the blood plasma and liver of fish (Table 3). Studies of individual RAs revealed their different capabilities of accumulating in tissues of living organisms. Oikari et al. [31] noted that abietane acids accumulated in fish tissues to the greatest extent, while pimarane acids, in much smaller amounts. Probably, this pattern is associated with lower bioavailability and solubility of pimarane-type acids. Blood-accumulated RAs enter the liver and pose toxic effects to organisms. There are literature data relevant to RA effects on the erythrocyte and hepatocyte energetics, using the example of rainbow trout (O. mykiss). DHA introduction causes a sharp increase in oxygen consumption and heat release, leading to ATP depletion. In RA-contaminated fish, hepatitis is generally observed, resulting from the erythrocyte hemolysis and hepatocyte damage [6]. A similar effect has been reported for cells in the central nervous system of fish. For instance, the addition of DHA provokes increased oxygen consumption, a decreased ATP level, and promotes Ca2+ release from intracellular stores [5].
Table 3.
RA bioaccumulation in fish organs.
| Study Object | Concentration | Conditions | Reference |
|---|---|---|---|
| Fine flounder Paralychthys adspersus Small-eyes flounder P. microps | |||
| Bile | 30.5–41.9 µg/g, total RA content | Caught near the PPM effluent discharge site | [4] |
| Rainbow trout O. mykiss | |||
| Bile | <200 µg/g DHA | After 57 days of exposure to PPM effluents | [32] |
| Blood plasma | 155–318 µg/g DHA | After 4 days of exposure to DHA (1.2 mg/L) in water | [31] |
| Liver | 98–103 µg/g DHA | After 4 days of exposure to DHA (1.2 mg/L) | |
| 202–351 µg/g, total RA content | After 2 days of exposure to a mixture of RAs (1.4 mg/L) in water | ||
| Kidney | 47–114 µg/g DHA | After 4 days of exposure to DHA (1.2 mg/L) | |
| 72–115 µg/g, total RA content | After 2 days of exposure to a mixture of RAs (1.4 mg/L) in water | ||
Widely spread in river and sea reservoirs, RAs are recorded (up to 8 µg/m3) as an inhalable particulate in the air at lumber mill work areas [33]. Analysis of smoke from the combustion of coniferous wood revealed DHA, ABA, PA, and IPA, as well as their hydroxy- and oxo-derivatives in suspended fine (2.5 µm) solids. At the same time, of all the substances (aliphatic and aromatic hydrocarbons, alcohols, acids, and phenols) detected, DHA was present in the greatest (23.8 µg/g of an extract) concentration [34].
There are data on RA migration from rosin, paper, and cardboard into food [35,36,37]. Currently, rosin consisting of more than 70% of RAs is used in dentistry as a component of periodontal dressings and cements, as well as a root canal filling [8]. Importantly, many RAs are resistant to environmental factors. For example, DHA has been preserved in pine bark compost for 12 months [38].
Ubiquitously distributed, RAs can influence both the aquatic fauna and humans. A mixture of individual RAs is characterized by high (concentration-dependent) cytotoxicity to human epithelial cells, polymorphonuclear leukocytes, and gingival fibroblasts [7,8]. Long-term exposure to RAs can provoke asthma and chronic pulmonary diseases in employees at wood-processing industries [39]. AA and DHA extracted from food packaging paper in high concentrations can promote a tumor formation [9]. Despite their obvious toxicity on the human body, some RAs can find potential applications in drug development, since they have anti-atherosclerotic [40], anti-inflammatory [24], antidiabetic [41,42], antitumor [43], osteoprotective [44], cytotoxic [45], antimicrobial [12], and anti-biofilm [46] actions.
3. Biodegradation of RAs
Biological treatment of PPM effluents is traditionally based on methods successfully employed in clean-up procedures of domestic wastewater. Biotreatments are classified into aerobic and anaerobic. Aerobic processes using aerated lagoons and activated sludge systems are the main options for complete degradation of organic material to CO2 and H2O or its transformation into eco-friendly compounds by natural microorganisms [47]. According to some authors [48,49], activated sludge and aerated lagoon treatments can reduce the level of abietane-type RAs in PPM effluents by 90%. However, removal of pimarane-type RAs is less efficient and does not exceed 60%.
It is generally accepted that most RAs can be removed by the aforementioned bioremediation systems. However, changes in the effluent composition, the properties of the degradation medium, the nutrient availability, and the state of the microbial community affect the treatment system efficiency and, in some cases, can lead to the release of more toxic and persistent compounds into the environment [14,50]. To prevent such disturbances, it is necessary to study in detail the composition of a microbial community and the role of each individual microbial species in biological treatment systems.
Because RAs are widespread in nature, microorganisms capable of degrading these ecotoxicants were discovered in various samples from river reservoirs [51], biological treatment systems [52,53], forest, agricultural, and Arctic soils [54,55], and soil and water contaminated with petroleum products [56]. Since the 1990s, a considerable number of pure bacterial and fungal cultures of RA degraders have been isolated (Table 4). The majority of aerobic bacterial isolates use RAs as a sole carbon source; still, there are data on bacteria that transform RAs but do not grow on them. Such bacterial cultures include proteobacteria isolated from compost by a group of Canadian scientists [57] and actinobacteria isolated from oil-contaminated soil and water bodies [51,56]. Fungal cultures usually transform RAs into hydroxy derivatives without using RAs as a source of carbon. The biodiversity of microbial biodegraders is basically represented by Alpha-, Beta-, Gamma-, and Deltaproteobacteria (Table 4). Data on the use of gram-positive bacteria for RA biodegradation are still scarce. The work has mainly been done employing actinobacteria and bacilli.
Table 4.
Microorganisms capable of RA biodegradation.
| Strain | Substrate | Reference |
|---|---|---|
| Gram-negative | ||
| Alcaligenes sp. D11-13 | DHA | [53] |
| Betaproteobacterium sp. DhA-71, DhA-73 | DHA | [57] |
| Burkholderia cepacia F45L5 | DHA, ABA, IPA | [58] |
| Burkholderia sp. DhA-54 | DHA | [59] |
| Burkholderia sp. IpA-51 | IPA | [59] |
| B. xenovorans LB400 | DHA, ABA, PA | [60,61,62] |
| Pseudomonas abietaniphila BKME-9 | DHA, ABA | [52,63] |
| P. fluorescens NRRL B21432 | Mixture of RAs | [64] |
| P. marginalis E-001624 | Mixture of RAs | [65] |
| P. mohnii IpA-2T, P. moorei RW10T | IPA | [66] |
| “Pseudomonas multiresinivorans” * (P. nitroducent) IpA-1 * |
IPA | [67] |
| P. reinekei Mt1 | IPA | [66] |
| Pseudomonas sp. A19-6a | ABA | [53] |
| Pseudomonas sp. DhA-92 | DHA | [55] |
| Pseudomonas sp. IpA-2 | IPA | [67] |
| Pseudomonas sp. IpA-93, IpA-95 | IPA | [55] |
| P. vancouverensis Dha-51 | DHA | [59] |
| Ralstonia sp. BKME-6 | DHA | [52] |
| Serratia marcescens NRRL B21429 | Mixture of RAs | [64] |
| Sphingomonas sp. DhA-33 | DHA | [54,68] |
| Sphingomonas sp. DhA-95 | DHA | [55] |
| Xanthomonas campestris NRRL B21430 | Mixture of RAs | [64] |
| Zoogloea ramigera DhA-35 | DHA | [68] |
| Gram-positive | ||
| Bacillus psychrophilus | DHA | [69] |
| Dietzia maris IEGM 55T | DHA | [56] |
| Gordonia rubripertincta IEGM 104, IEGM 105, IEGM 109 | DHA | [51] |
| G. terrae IEGM 150 | DHA | [51] |
| Mycobacterium sp. DhA-55 | DHA | [54] |
| Mycobacterium sp. IpA-13 | IPA | [67] |
| Rhodococcus erythropolis IEGM 267 | DHA | [51] |
| R. rhodochrous IEGM 107 | DHA | [51] |
| R. ruber IEGM 80 | DHA | [51] |
Note: * The former name of the bacterial taxon is in upright font and in quotation marks. The current name of the taxon is given in parenthesis.
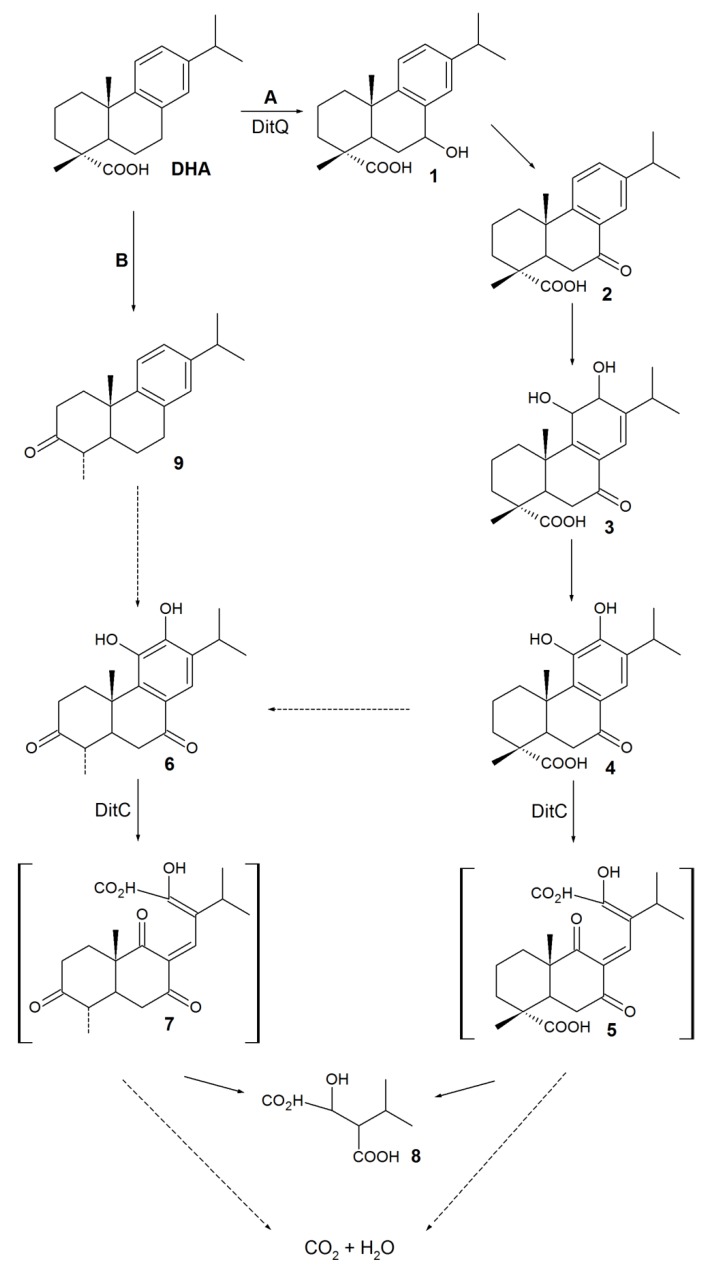
A general DHA biodegradation pathway was proposed based on genetic investigation of bacterial strains capable of degrading the abietane RAs isolated from a PPM (Scheme 1) [70,71,72].
Scheme 1.
The proposed pathway for bacterial DHA degradation.
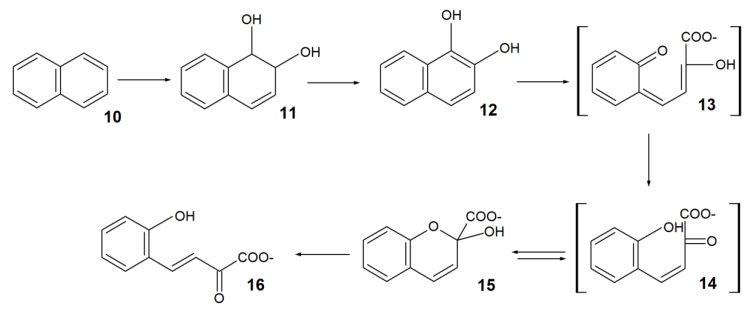
Presumably, the first step of biodegradation includes C-7 hydroxylationof DHA (Scheme 1, pathway A) or C-3 oxidation followed by decarboxylation to 3-oxo-dehydroabietin 9 (Scheme 1, pathway B). Compounds 1 and 9 were detected simaultaneously, which did not allow judging on the direction of aprimary oxidative reaction at C-3 and C-7 positions. In this regard, two alternative pathways for the intermediate 6 formation were suggested. The discovery of the DitA dioxygenase complex from P. abietaniphila BKME-9 [70] catalyzing the formation of 7-oxo-11,12-dihydroxy-8,13-abietadienic acid 3 from 7-oxo-derivative 2 gives evidence in favor of pathway A. Aromatization of diol 3 via 11,12-dehydration is supposed to lead to 7-oxo-11,12-diol 4, and the latter is further oxidized and decarboxylated at C-3 to form an intermediate 6. Identification of P. abietaniphila BKME-9 dioxygenase involved in the meta-cleavage of diterpenoids and detection of 2-isopropylmalic acid 8 entail that diols 4 and 6 could be further degraded via the aromatic ring cleavage resulting in the formation of possible intermediates 5 and 7 [15,71].The authors [15] believe that a general pathway of DHA biodegradation is somewhat similar to the initial pathways of bacterial degradation of polycyclic aromatic hydrocarbons (PAHs) (Scheme 2).
Scheme 2.
Bacterial degradation of polycyclic aromatic hydrocarbons (PAHs) on the example of naphthalene.
Using naphthalene 10 asan example, the authors [73] showed that a bacterial dioxygenase first catalyzed the formation of dihydrodiol 11 and then its aromatization, resulting in 1,2-dihydroxynaphthalene 12. Subsequent cleavage of diol 12 occurred via the formation of unstable compounds 13, 14, and hemiacetal 15. The latter, spontaneously or enzymatically, was isomerized to trans-o-hydroxybenzylidenepyruvate 16. Assuming that DHA biodegradation proceeds similar to that of PAHs—via hemiacetal formation—it becomes clear why compounds 5 and 7 are not detected among biodegradation products [15].
Cheremnykh et al. [51] employed R. rhodochrous IEGM 107 and reported that metabolites of DHA were detected in the post-cultural medium when additional carbon and energy (n-hexadecane) sources were used. The mass spectra of metabolites corresponded to the known spectra of 7-oxo-DHA 2 and 7-oxo-11,12-dihydroxy-8,13-abietadienic acid 3 characteristic of P. abietaniphila BKME-9 [70]. The process of actinobacterial degradation of DHA by R. rhodochrous IEGM 107 cells seems to proceed likewise via C-7 oxidation followed by dihydroxylation and further meta-cleavage of the aromatic ring.
The formation of macroaggregates, the changes in morphometric parameters (an increased cell size) and the cell surface properties (an increased mean-square roughness, a decreased electrokinetic potential) of DHA-exposed actinobacteria were shown using different microscopic methods (namely, phase contrast, atomic force, and confocal laser scanning) and measurements of the cell electrokinetic potential [51]. The identified changes have been considered as mechanisms of actinobacterial adaptation to DHA exposure and, consequently, of their resistance to the DHA toxic effect.
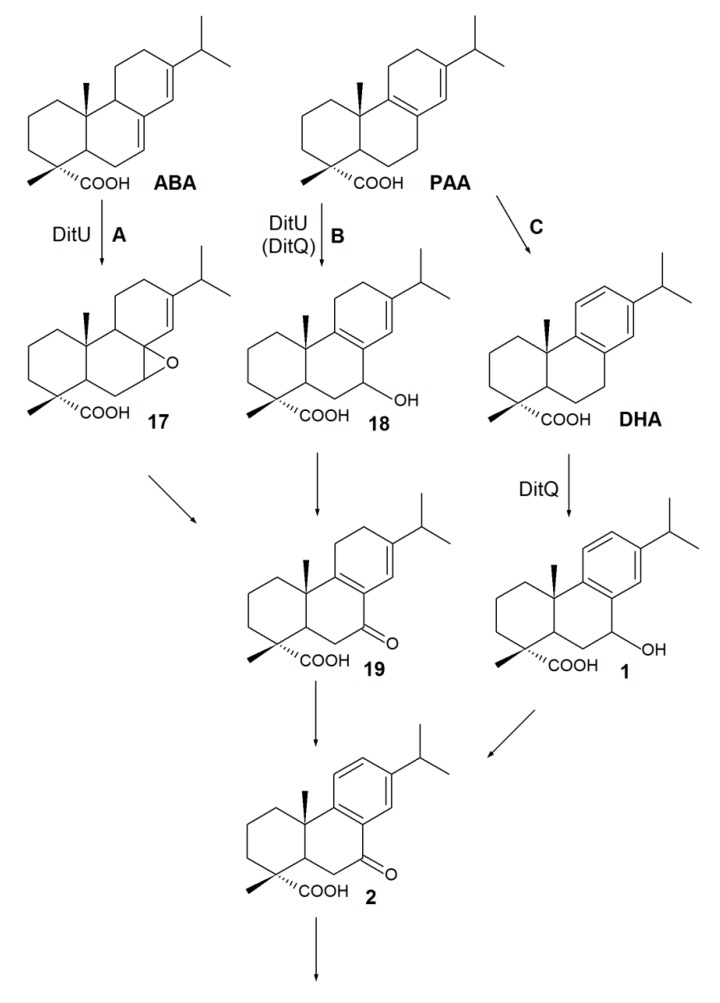
Since the early 1990s, studies of the substrate specificities of RA-degrading bacteria have shown that the abilities of isolates to use DHA as the sole carbon source are in agreement with their ability to grow on any abietane-type derivatives [52,68]. The abietane-type RAs were previously thought to be metabolized via DHA [70]. However, using P. abietaniphila BKME-9 mutant strains, it was later shown that a mutation in gene ditQ limits pseudomonades’ growth in the presence of DHA but has little effect on the degradation of non-aromatic acids, like ABA and PA [60]. A genetic study of the biodegrading strains—P. abietaniphila BKME-9 [70,71] and B. xenovorans LB400 [62]—suggested a common convergent pathway for abietane-type RA biodegradation, resulting in the common intermediate7-oxo-DHA 2 (Scheme 3).
Scheme 3.
The proposed convergent pathway for abietane-type RA biodegradation.
Presumably, the first stage of ABA biodegradation consists of oxidation to a 7,8-epoxy-ABA intermediate 17 (Scheme 3, pathway A), and the opening and rearrangement of an oxirane ring, leading to 7-oxo-PAA 19 formation [60,61,62,70,71]. The proposed mechanism (Scheme 3) explains the detection of compound 19 in the culture medium of B. xenovorans LB400 during ABA degradation [62]. Accumulation of 7-oxo-DHA 2 in the ABA degradation medium was probably due to aromatization of 7-oxo-PAA 19 to 7-oxo-DHA 2 [62]. Mineralization of PAA proceeded through two alternative pathways: via C-7 hydroxylation (compound 18) followed by 7-oxo-derivate 19 formation (Scheme 3, pathway B) or through the key stage of aromatization to DHA (Scheme 3, pathway C) [60,61,62,70,71]. Pathway B is in agreement with the results reported by Smith et al. [62]. They detected the formation of 7-oxo-PAA19 in the B. xenovorans LB400 culture medium supplemented with PAA. Thus, 7-oxo-PAA19 is a “crossing point” of ABA and PAA metabolic pathways, while 7-oxo-DHA 2 is a “crossing point” of all abietane-type RAs.
The data on biodegradation pathways for pimarane-type RAs are sporadic. The aerobic gram-negative bacteria—known to grow on abietanes—cannot use pimarane acids as the sole carbon source. However, isolates growing on pimaranes can use both pimarane and abietane acids [52,67,68]. Present or absent in the molecules of these compounds, an isopropyl group is pivotal in dictating the RA biodegradation pathways. It was shown in [67] that isolates of Pseudomonas sp. IpA-1 and IpA-2, obtained from an enriched culture and grown in the presence of IPA, exhibited different degrading activities against abietanes. Pseudomonas sp. IpA-1 required IPA in the culture medium to effectively use abietanes, while the strain IpA-2 used abietanes as the sole carbon source.
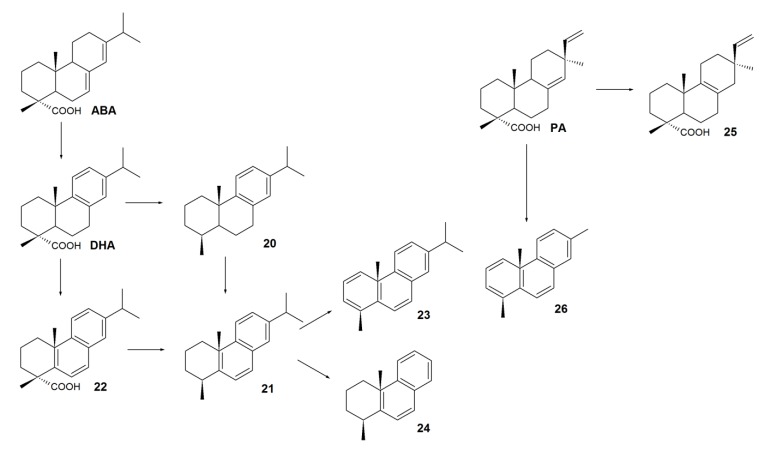
The aforementioned degradation examples of various RAs have been mainly described for aerobic microorganisms. Under oxygen-free conditions, RAs could be biologically transformed; however, there is no convincing evidence of complete degradation of their carbon skeletons. Mohn et al. [54] showed that RAs are difficult to degrade under oxygen-free conditions. Moreover, pure anaerobic cultures capable of using RAs as the sole carbon source have not yet been isolated. Despite considerable difficulties associated with RA degradation under anoxic conditions, a group of New Zealand scientists led by Dr. Tavendale [74,75] carried out a large-scale study of DHA, ABA, and PA biotransformations under anoxic conditions into neutral derivatives and described for the first time new pathways of anoxic conversion of RAs in 1997. After 264 days of an anaerobic sediment incubation containing deuterium-labeled RAs, several compounds were isolated, suggesting a pathway for anaerobic metabolism of ABA, DHA, and PA (Scheme 4). 18-Norabietatrien 20 and tetrahydroretene 21 were registered as major products of anaerobic DHA degradation. Presumably, tetrahydroretene 21 was formed by the alternative pathways: either via ring B aromatization of 18-norabietatrien 20 or via carboxylation of abieta-5(10),6,8,11,13-pentaen-18-oic acid 22. A small amount of tetrahydroretene 21 was transformed into retene 23 and 1-methyltetrahydrophenantrene 24. Degradation of ABA under anaerobic conditions was also observed. However, the nature of this process is questionable (either biotic or abiotic) because the decreases in ABA concentration in the experiment and in the control were similar [74,75]. Degradation of pimarane-type RAs under anoxic conditions is still unclear A slight increase in pimar-8-en-18-oic acid 25 concentration was detected [15]. It is likely that anaerobic biocatalysis of pimarane acids proceeds similarly to that of abietane acids in a multi-stage process with the pimanthrene 26 formed [74,75]. Remarkably, the large-scale processes of anaerobic treatment are not widely applied for RA removal from effluents. Apart from being time-consuming, the employment of closed systems leads to concentrating of effluents. In this case, high concentrations of RAs inhibit the enzyme systems of anaerobic microorganisms [14,76].
Scheme 4.
Proposed anaerobic pathways for ABA, DHA, and PA degradation.
4. Biotransformation of RAs to Bioactive Compounds
Up to date, RA-derived compounds with different pharmacological effects—anti-inflammatory [77], antimicrobial [12,78], fungicidal [12,22,23,79], anxiolytic [80], antiviral [23], antitumor [23,43,81], and anti-angiogenic [82]—have been described. In addition, RA derivatives can be used as intermediates in the synthesis of bioactive compounds [13,83] and pharmaceuticals [84,85]. To obtain novel compounds with biological properties, different techniques are used, particularly a chemical transformation [86,87]. Methods of chemical synthesis, however, often require expensive catalysts and introduction of protecting groups of reactive functional centers of the molecules. Widely known chemical conversions of RAs usually include classical transformations at rings B and C [83] because it is very difficult to perform regio- and stereoselective reactions at ring A using chemical methods. An alternative approach to structural modifications of natural compounds—particularly RAs—is microbial biotransformation, which does not require aggressive chemicals, proceeds in one technological stage, and is highly regio- and stereoselective [88,89]. Using inhibitors of enzyme systems allows for some intermediates of the microbial RA degradation with significant biological activities. For example, 7β-hydroxy-DHA 1 (Table 5)—a frequently registered product of DHA biotransformation—has antimicrobial, fungicidal, and selective antitumor properties [90,91]. Directed microbial transformations provide novel products unusual for the RA biodegradation pathways previously proposed.
Table 5.
Transformation products of abietane-type RAs using fungi, and their bioactivities.
| Compound | Biological Activity | Biocatalyst | Reference |
|---|---|---|---|
| 1β-hydroxy-DHA (27) | Antimicrobial, inhibitory activity against α-glucosidase |
Aspergillus niger, Cephalosporium aphidicola, Cunninghamella elegans, Fusarium moniliforme, F. oxysporum, Gibberella fujikuroi, Neurospora crassa, Phlebiopsis gigantea, Rhizopus stolonifera |
[22,90,91,92,93] |
| 2α-hydroxy-DHA (28) | Antimicrobial, selective antitumor | Mucor ramannianus | [91] |
| 7β-hydroxy-DHA (1) | Antimicrobial, fungicidal, antitumor | A. niger, N. crassa | [78,79,90,91,94] |
| 15-hydroxy-DHA (29) | Anti-inflammatory. An intermediate of antiviral and antitumor agent synthesis |
C. aphidicola, C. elegans, G. fujkuroi, R. stolonifera |
[22,83] |
| 16-hydroxy-DHA (30) | Antimicrobial |
C. aphidicola, C. elegans, G. fujkuroi, R. stolonifera |
[22] |
Numerous abietane-type RA bioconversions using fungal strains have been reported. Basically, fungi modify a substrate molecule by stereoselective hydroxylation. Hydroxyl groups can be introduced at various positions of the molecule. In the case of RAs, hydroxylation reactions have been most frequently registered at C-1, C-2, C-7, C-15, and C-16 carbon atoms. DHA and ABA are usually used as substrates for directed transformation using fungi. Table 5 shows monohydroxy derivatives with biological activities obtained using fungal cultures.
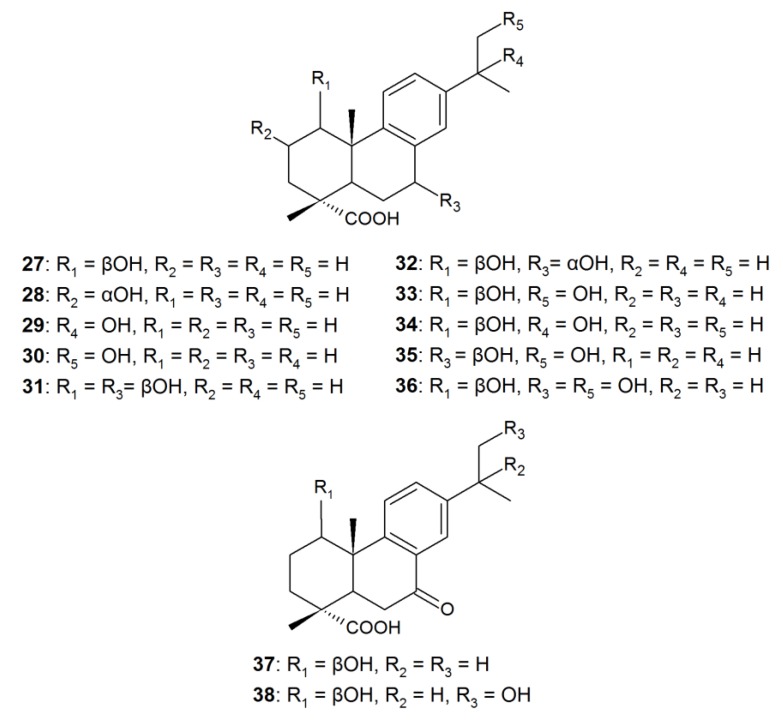
In addition, hydroxylation of DHA can proceed selectively or lead to the accumulation of several hydroxy derivatives of DHA. For example, when cultures of C. elegans TSY 0865, R. stolonifer ATCC-10404, G. fujikuroi ATCC-10704, and C. aphidicola IMI-68689 were used, three regioisomeric monohydroxy derivatives of DHA—acids 27, 29, and 30—exhibiting an antibacterial effect and an inhibitory activity against α-glucosidase were the biotransformation products (Figure 2) [22]. Fungi are capable of transforming RAs into di- and trihydroxy derivatives at C-1, C-2, C-7, C-15, and C-16 positions. For example, in addition to 1β-hydroxy-DHA 27 and 7β-hydroxy-DHA 1, a 1β,7β-dihydroxy derivative 31 with an antimicrobial activity was registered among the products of DHA biotransformation using A. niger cells [90]. In case of P. gigantea, together with 1β-hydroxy-DHA 27, the hydroxy derivatives of DHA—1β,7α-dihydroxy 32, di- (33–35), and tri- (36) hydroxy derivatives—were observed, including those with a β-orientation of the hydroxyl group at C-7 [93]. Similar di- and trihydroxy DHA derivatives (33–36) were detected in the transformation medium of Trametes versicolor [93]. In addition to DHA hydroxylation by P. gigantea and T. versicolor cells, transformation of this acid to 7-oxo derivatives—1β-hydroxy-7-oxo-DHA 37 and 1β,16-dihydroxy-7-oxo-DHA 38—was observed (Figure 2) [93]. At the same time, examples of selective fungal transformation of pimarane-type RAs are few because PA and IPA are recalcitrant substrates for microorganisms.
Figure 2.
Structures of RA transformation products using fungi.
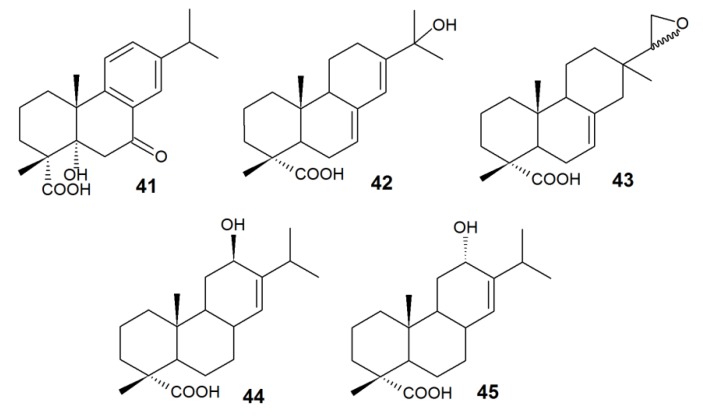
Directed biotransformations of abietane-type RAs by bacterial cells to compounds unusual for the above described biodegradation pathways (Scheme 1, Scheme 3) are less studied. For example, 7-oxo-2 and 5α-hydroxy-7-oxo-DHA 41 accumulated in the culture media for DHA transformations using Burkholderia sp., Cupriavius sp., and Pseudomonas sp. with a modified ditA1 gene [95].
New metabolites—5α-hydroxy-DHA 39 and 15,16,17-trinor-abietane-type compound 40—produced using R. erythropolis IEGM 267 cells pre-grown in the presence of DHA suggested a novel pathway of DHA biotransformation. Earlier, only single facts of hydroxylation at C-5 of the abietanes were described [95]. Biotransformation involving R. erythropolis IEGM 267 cells most likely occurs by oxidation of the parent compound molecule at C-5 of the carbon ring followed by deisopropylation of the aromatic ring (Scheme 5).
Scheme 5.
DHA biotransformation using R. erythropolis IEGM 267.
To degrade and transform RAs, enzymatic complexes as well as whole bacterial cells are often employed. The modified (by genetic transformation or gene cloning) cells of Escherichia coli with high enzyme efficiency are usually used to produce these complexes. Cytochrome-dependent bacterial enzymes CYP105A1 and CYP106A2 specific for RAs have been described in literature [25,96,97]. For example, CYP105A1 from Streptomyces griseolus catalyzed the formation of 15-hydroxy derivatives 29 and 42 from DHA and ABA, respectively, and 15,16-epoxy-IPA 43 from IPA [25]. 12-Hydroxy derivatives 44 and 45 were detected in the reaction medium containing ABA and CYP106A2 purified from B. megaterium cells (Figure 3) [96]. Enzymatic systems catalyze certain types of reactions, and this allows for the control of biotransformation processes. However, this method requires a significant investment of time and money as regards enzyme purification.
Figure 3.
Structures of RA biotransformation products using bacteria and purified bacterial enzymatic complexes.
5. Conclusions
The most promising means of RA neutralization is the application of methods employing the enzymatic activity of microorganisms. Among the described cultures capable of complete RA degradation, the most common are mycelial fungi and proteobacteria isolated from RA-polluted sites. Gram-positive RA-degrading bacteria are, however, represented by only a few strains belonging to the genera Bacillus, Mycobacterium, and Rhodococcus. In recent years, intensive studies of RA biodegradation processes have favored the description of possible pathways for RA bioconversion.
Since the 2000s, transformation of RAs tailored to produce bioactive compounds for biotechnology needs has become increasingly important. So far, derivatives with significant anti-inflammatory, antimicrobial, fungicidal, anxiolytic, antiviral, antitumor, and anti-angiogenic activities have been described. It is of relevance due to the lack of therapeutic agents in certain areas of medicine (cardiovascular, tumor diseases, and immune system diseases).
It is noteworthy that, despite many processes of biological RA degradation and transformation having been described, the majority have significant drawbacks. Bacterial cultures usually exhibit their activities at RA concentrations not higher than 250 mg/L, while in effluents they can be exposed to RA concentrations exceeding this value (up to 1500 mg/L). The use of fungi capable of catalyzing a wide range of reactions implies certain risks due to their seed (spore) material and the ability to synthesize mycotoxins. To date, a number of actinobacterial strains related to Dietzia maris, Gordonia rubripertincta, G. terrae, Rhodococcus erythropolis, R. rhodochrous, and R. ruber and capable of converting higher (up to 500 mg/L) DHA concentrations have been discovered. The experimental data presented in this review create the prerequisites for the implementation of advanced technology solutions for an effective removal of RAs from PPM effluents.
Funding
The work was funded by the Russian Foundation for Basic Research (grants No. 18-34-00109 and No. 18-29-05006), and the Russian Federation Ministry of Education and Science (State Assignments 01201353246 and 6.3330.2017/4.6).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Food and Agriculture Organization of The United Nations, (FAO) Forestry Production and Trade (the Database) [(accessed on 9 July 2019)]; Available online: http://www.fao.org/faostat/en/#data/FO/visualize.
- 2.Singh P., Srivastava N., Jagadish R., Upadhyay A. Effect of toxic pollutants from pulp and paper mill on water and soil quality and its remediation. Int. J. Lakes Rivers. 2019;12:1–20. [Google Scholar]
- 3.Ottavioli J., Paoli M., Casanova J., Tomi F., Bighelli A. Identification and quantitative determination of resin acids from corsican Pinus pinaster Aiton oleoresin using 13C-NMR spectroscopy. Chem. Biodivers. 2019;16:e1800482. doi: 10.1002/cbdv.201800482. [DOI] [PubMed] [Google Scholar]
- 4.Hernández V., Silva M., Gavilán J., Jiménez B., Barra R., Becerra J. Resin acids in bile samples from fish inhabiting marine waters affected by pulp mill effluents. J. Chil. Chem. Soc. 2008;53:1718–1721. doi: 10.4067/S0717-97072008000400018. [DOI] [Google Scholar]
- 5.Zheng J., Nicholson R. Action of resin acids in nerve ending fractions isolated from fish central nervous system. Environ. Toxicol. Chem. 1998;17:1852–1859. doi: 10.1002/etc.5620170928. [DOI] [Google Scholar]
- 6.Rissanen E., Krumschnabel G., Nikinmaa M. Dehydroabietic acid, a major component of wood industry effluents, interferes with cellular energetics in rainbow trout hepatocytes. Aquat. Toxicol. 2003;62:45–53. doi: 10.1016/S0166-445X(02)00066-8. [DOI] [PubMed] [Google Scholar]
- 7.Söderberg T.A., Johansson A., Gref R. Toxic effects of some conifer resin acids and tea tree oil on human epithelial and fibroblast cells. Toxicology. 1996;107:99–109. doi: 10.1016/0300-483X(95)03242-8. [DOI] [PubMed] [Google Scholar]
- 8.Sunzel B., Söderberg T.A., Johansson A., Hallmans G., Gref R. The protective effect of zinc on rosin and resin acid toxicity in human polymorphonuclear leukocytes and human gingival fibroblasts in vitro. J. Biomed. Mater. Res. 1997;37:20–28. doi: 10.1002/(SICI)1097-4636(199710)37:1<20::AID-JBM3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 9.Ohmori K., Kawamura Y. Cell transformation activities of abietic acid and dehydroabietic acid: Safety assessment of possible contaminants in paper and paperboard for food contact use. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2009;26:568–573. doi: 10.1080/02652030802471221. [DOI] [PubMed] [Google Scholar]
- 10.Nestmann E.R., Lee E.G., Mueller J.C., Douglas G.R. Mutagenicity of resin acids identified in pulp and paper mill effluents using the salmonella/mammalian-microsome assay. Environ. Mutagen. 1979;1:361–369. doi: 10.1002/em.2860010408. [DOI] [PubMed] [Google Scholar]
- 11.Kinae N., Hashizume T., Makita T., Tomita I., Kimura I., Kanamori H. Studies on the toxicity of pulp and paper mill effluents—I. Mutagenicity of the sediment samples derived from kraft paper mills. Water Res. 1981;15:17–24. doi: 10.1016/0043-1354(81)90177-9. [DOI] [Google Scholar]
- 12.Burčová Z., Kreps F., Greifová M., Jablonský M., Ház A., Schmidt Š., Šurina I. Antibacterial and antifungal activity of phytosterols and methyl dehydroabietate of Norway spruce bark extracts. J. Biotechnol. 2018;282:18–24. doi: 10.1016/j.jbiotec.2018.06.340. [DOI] [PubMed] [Google Scholar]
- 13.Miyajima Y., Saito Y., Takeya M., Goto M., Nakagawa-Goto K. Synthesis of 4-epi-parviflorons A, C, and E: Structure-activity relationship study of antiproliferative abietane derivatives. J. Org. Chem. 2019;84:3239–3248. doi: 10.1021/acs.joc.8b02832. [DOI] [PubMed] [Google Scholar]
- 14.Liss S.N., Bicho P.A., Saddler J.N. Microbiology and biodegradation of resin acids in pulp mill effluents: A minireview. Can. J. Microbiol. 1997;75:599–611. doi: 10.1139/m97-086. [DOI] [PubMed] [Google Scholar]
- 15.Martin V.J.J., Yu Z., Mohn W.W. Recent advances in understanding resin acid biodegradation: Microbial diversity and metabolism. Arch. Microbiol. 1999;172:131–138. doi: 10.1007/s002030050752. [DOI] [PubMed] [Google Scholar]
- 16.Keeling C.I., Bohlmann J. Diterpene resin acids in conifers. Phytochemistry. 2006;67:2415–2423. doi: 10.1016/j.phytochem.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Rogachev A.D., Salakhutdinov N.F. Chemical composition of Pinus sibirica (Pinaceae) Chem. Biodivers. 2015;12:1–53. doi: 10.1002/cbdv.201300195. [DOI] [PubMed] [Google Scholar]
- 18.Gonçalves M.D., Bortoleti B.T.S., Tomiotto-Pellissier F., Miranda-Sapla M.M., Assolini J.P., Carloto A.C.M., Carvalho P.G.C., Tudisco E.T., Urbano A., Ambrósio S.R., et al. Dehydroabietic acid isolated from Pinus elliottii exerts in vitro antileishmanial action by pro-oxidant effect, inducing ROS production in promastigote and downregulating Nrf2/ferritin expression in amastigote forms of Leishmania amazonensis. Fitoterapia. 2018;128:224–232. doi: 10.1016/j.fitote.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 19.Simoneit B.R.T., Cox R.E., Oros D.R., Otto A. Terpenoid compositions of resins from Callitris species (Cupressaceae) Molecules. 2018;23:3384. doi: 10.3390/molecules23123384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schicchi R., Geraci A., Rosselli S., Spinella A., Maggio A., Bruno M. Phytochemical investigation of the needles of Abies nebrodensis (Lojac.) Mattei. Nat. Prod. Res. 2019:1–6. doi: 10.1080/14786419.2019.1576044. [DOI] [PubMed] [Google Scholar]
- 21.Wajs-Bonikowska A., Smeds A., Willför S. Chemical composition and content of lipophilic seed extractives of some Abies and Picea species. Chem. Biodivers. 2016;13:1194–1201. doi: 10.1002/cbdv.201600014. [DOI] [PubMed] [Google Scholar]
- 22.Choudhary M.I., Atif M., Shan S.A.A., Sultan S., Erum S., Khan S.N. Atta-Ur-Rahman Biotransformation of dehydroabietic acid with microbial cell cultures and α-glucosidase inhibitory activity of resulting metabolites. Int. J. Pharm. Pharm. Sci. 2014;6:375–378. [Google Scholar]
- 23.González M.A., Perez-Guaita D., Correa-Royero J., Zapata B., Agudelo L., Mesa-Arango A., Betancur-Galvis L. Synthesis and biological evaluation of dehydroabietic acid derivatives. Eur. J. Med. Chem. 2010;45:811–816. doi: 10.1016/j.ejmech.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Kang M.S., Hirai S., Goto T., Kuroyanagi K., Lee J.Y., Uemura T., Ezaki Y., Takahashi N., Kawada T. Dehydroabietic acid, a phytochemical, acts as ligand for PPARs in macrophages and adipocytes to regulate inflammation. Biochem. Biophys. Res. Commun. 2008;369:333–338. doi: 10.1016/j.bbrc.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Janocha S. Ph.D. Thesis. Univarsität des Saarlandes; Saarbrücken, Germany: 2013. Umsatz von Harzsäuren Durch die Bakteriellen Cytochrome CYP105A1 und CYP106A2—Strukturelle Grundlagen und Potentielle Anwendungen. [Google Scholar]
- 26.Plemenkov V.V., Appolonova S.A., Kirlitsa D.A. To the question of the native content of resin acids in the galipot of conifers. Chem. Comput. Simulation. Butl. Commun. 2004;5:30–32. [Google Scholar]
- 27.Peng G., Roberts J.C. Solubility and toxicity of resin acids. Wat. Res. 2000;34:2779–2785. doi: 10.1016/S0043-1354(99)00406-6. [DOI] [Google Scholar]
- 28.Zanella E. Effect of pH on acute toxicity of dehydroabietic acid and chlorinated dehydroabietic acid to fish and Daphnia. Bull. Environ. Contam. Toxicol. 1983;30:133–140. doi: 10.1007/BF01610111. [DOI] [PubMed] [Google Scholar]
- 29.Volkman J.K., Holdsworth D.G., Richardson D.E. Determination of resin acids by gas chromatography and high-perfomance liquid chromatography in paper mill effluent, river waters and sediments from the upper Derwent Estuary, Tasmania. J. Chromatogr. 1993;643:209–219. doi: 10.1016/0021-9673(93)80555-M. [DOI] [Google Scholar]
- 30.Leppanen H., Marttinen S., Oikari A. The use of fish bile metabolite analyses as exposure biomarkers to pulp and paper mill effluents. Chemosphere. 1998;36:2621–2634. doi: 10.1016/S0045-6535(97)10226-0. [DOI] [Google Scholar]
- 31.Oikari A., Holmbom B., Bister H. Uptake of resin acids into tissues of trout (Salmo gairdneri Richardson) Ann. Zool. Fenn. 1982;19:61–64. [Google Scholar]
- 32.Lindesjöö E., Adolfsson-Erici M., Ericson G., Förlin L. Biomarker responses and resin acids in fish chronically exposed to effluents from a total chlorine-free pulp mill during regular production. Ecotoxicol. Environ. Saf. 2002;53:238–247. doi: 10.1006/eesa.2002.2211. [DOI] [PubMed] [Google Scholar]
- 33.Teschke K., Demers P.A., Davies H.W., Kennedy S.M., Marion S.A., Leung V. Determinants of exposure to inhalable particulate, wood dust, resin acids, and monoterpenes in a lumber mill environment. Ann. Occup. Hyg. 1999;43:247–255. doi: 10.1016/S0003-4878(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 34.Simoneit B.R.T., Rogge W.F., Lang Q., Ja R. Molecular characterization of smoke from campfire burning of pine wood (Pinus elliottii) Chemosphere. 2000;2:107–122. doi: 10.1016/S1465-9972(99)00048-3. [DOI] [Google Scholar]
- 35.Ozaki A., Ooshima T., Mori Y. Migration of dehydroabietic and abietic acids from paper and paperboard food packaging into food-simulating solvents and Tenax TA. Food Addit. Contam. 2006;23:854–860. doi: 10.1080/02652030600743813. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y., Zhang S., Geng Z., Wang D., Liu F., Zhang M. Analysis of abietic acid and dehydroabietic acid residues in raw ducks and cooked ducks. Poult. Sci. 2014;93:1–5. doi: 10.3382/ps.2014-04045. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y., Zhang S., Geng Z., Wang D., Liu F., Zhang M., Bian H., Xu W. Simultaneous determination of abietic acid and dehydroabietic acid residues in duck meat by HPLC-PAD-FLD. Food Anal. Methods. 2014;7:1627–1633. doi: 10.1007/s12161-014-9798-6. [DOI] [Google Scholar]
- 38.Tarabanko V.E., Ulyanova O.A., Kalacheva G.S. Investigation of dynamics of the content of terpene compounds in pine bark-based composts and their growth-promoting activity. Russ. J. Bioorganic Chem. 2010;1:121–126. [Google Scholar]
- 39.Ayars G.H., Altman L.C., Frazier E., Chi E.Y. The toxicity of constituents of cedar and pine woods pulmonary epitheii. J. Allergy Clin. Immunol. 1989;83:610–618. doi: 10.1016/0091-6749(89)90073-0. [DOI] [PubMed] [Google Scholar]
- 40.Suh S.J., Kwak C.H., Chung T.W., Park S.J., Cheeeei M., Park S.S., Seo C.S., Son J.K., Chang Y.C., Park Y.G., et al. Pimaric acid from Aralia cordata has an inhibitory effect on TNF-α-induced MMP-9 production and HASMC migration via down-regulated NF-κB and AP-1. Chem. Biol. Interact. 2012;199:112–119. doi: 10.1016/j.cbi.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Kang M.-S., Hirai S., Goto T., Kuroyanagi K., Kim Y.-I., Ohyama K., Uemura T., Lee J.-Y., Sakamoto T., Ezaki Y., et al. Dehydroabietic acid, a diterpene, improves diabetes and hyperlipidemia in obese diabetic KK-Ay mice. BioFactors. 2009;35:442–448. doi: 10.1002/biof.58. [DOI] [PubMed] [Google Scholar]
- 42.Hjortness M.K., Riccardi L., Hongdusit A., Ruppe A., Zhao M., Kim E.Y., Zwart P.H., Sankaran B., Arthanari H., Sousa M.C., et al. Abietane-type diterpenoids inhibit protein tyrosine phosphatases by stabilizing an inactive enzyme conformation. Biochemistry. 2018;57:5886–5896. doi: 10.1021/acs.biochem.8b00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X.C., Wang M., Pan Y.M., Yao G.Y., Wang H.S., Tian X.Y., Qin J.K., Zhang Y. Synthesis and antitumor activities of novel thiourea α-aminophosphonates from dehydroabietic acid. Eur. J. Med. Chem. 2013;69:508–520. doi: 10.1016/j.ejmech.2013.08.055. [DOI] [PubMed] [Google Scholar]
- 44.Thummuri D., Guntuku L., Challa V.S., Ramavat R.N., Naidu V.G.M. Abietic acid attenuates RANKL induced osteoclastogenesis and inflammation associated osteolysis by inhibiting the NF-KB and MAPK signaling. J. Cell. Physiol. 2018;234:443–453. doi: 10.1002/jcp.26575. [DOI] [PubMed] [Google Scholar]
- 45.Cao Q., Wang S.X., Cheng Y.X. Abietane diterpenoids with potent cytotoxic activities from the resins of Populus euphratica. Nat. Prod. Commun. 2019;14:1–5. doi: 10.1177/1934578X19850029. [DOI] [Google Scholar]
- 46.Fallarero A., Skogman M., Kujala J., Rajaratnam M., Moreira V.M., Yli-Kauhaluoma J., Vuorela P. (+)-Dehydroabietic acid, an abietane-type diterpene, inhibits Staphylococcus aureus biofilms in vitro. Int. J. Mol. Sci. 2013;14:12054–12072. doi: 10.3390/ijms140612054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kostamo A., Kukkonen J.V.K. Removal of resin acids and sterols from pulp mill effluents by activated sludge treatment. Water Res. 2003;37:2813–2820. doi: 10.1016/S0043-1354(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 48.Stuthridge T.R., Campin D.N., Langdon A.G., Mackie K.L., McFarlane P.N., Wilkins A.L. Treatability of bleached kraft pulp and paper mill wastewaters in a New Zealand aerated lagoon treatment system. Water Sci. Technol. 1991;24:309–317. doi: 10.2166/wst.1991.0487. [DOI] [Google Scholar]
- 49.Belmonte M., Xavier C., Decap J., Martinez M., Sierra-Alvarez R., Vidal G. Improved aerobic biodegradation of abietic acid in ECF bleached kraft mill effluent due to biomass adaptation. J. Hazard. Mater. 2006;135:256–263. doi: 10.1016/j.jhazmat.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 50.Werker A.G., Hall E.R. Limitations for biological removal of resin acids from pulp mill effluent. Water Sci. Technol. 1999;40:281–288. doi: 10.2166/wst.1999.0723. [DOI] [Google Scholar]
- 51.Cheremnykh K.M., Luchnikova N.A., Grishko V.V., Ivshina I.B. Bioconversion of ecotoxic dehydroabietic acid using Rhodococcus actinobacteria. J. Hazard. Mater. 2018;346:103–112. doi: 10.1016/j.jhazmat.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 52.Bicho P.A., Martin V., Saddler J.N. Growth, induction, and substrate-specificity of dehydroabietic acid-degrading bacteria isolated from a kraft mill effluent enrichment. Appl. Environ. Microbiol. 1995;61:3245–3250. doi: 10.1128/aem.61.9.3245-3250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan C.A., Wyndham R.C. Isolation and characterization of resin acid degrading bacteria found in effluent from a bleached kraft pulp mill. Can. J. Microbiol. 1996;42:423–430. doi: 10.1139/m96-058. [DOI] [PubMed] [Google Scholar]
- 54.Mohn W.W., Wilson A.E., Bicho P., Moore E.R. Physiological and phylogenetic diversity of bacteria growing on resin acids. Syst. Appl. Microbiol. 1999;22:68–78. doi: 10.1016/S0723-2020(99)80029-0. [DOI] [PubMed] [Google Scholar]
- 55.Yu Z., Stewart G.R., Mohn W.W. Apparent contradiction: Psychrotolerant bacteria from hydrocarbon-contaminated Arctic tundra soils that degrade diterpenoids synthesized by trees. Appl. Environ. Microbiol. 2000;66:5148–5154. doi: 10.1128/AEM.66.12.5148-5154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheremnykh K.M., Grishko V.V., Ivshina I.B. Bacterial degradation of ecotoxic dehydroabietic acid. Catal. Ind. 2017;9:331–338. doi: 10.1134/S207005041704002X. [DOI] [Google Scholar]
- 57.Yu Z., Mohn W.W. Isolation and characterization of thermophilic bacteria capable of degrading dehydroabietic acid. Can. J. Microbiol. 1999;45:513–519. doi: 10.1139/w99-028. [DOI] [PubMed] [Google Scholar]
- 58.Lindberg L.E., Holmbom B.R., Väisänen O.M., Weber A.M.-L., Salkinoja-Salonen M.S. Degradation of paper mill water components in laboratory tests with pure cultures of bacteria. Biodegradation. 2001;12:141–148. doi: 10.1023/A:1013175421662. [DOI] [PubMed] [Google Scholar]
- 59.Mohn W.W., Yu Z., Moore E.R.B., Muttray A.F. Lessons learned from Sphingomonas species that degrade abietane triterpenoids. J. Ind. Microbiol. Biotechnol. 1999;23:374–379. doi: 10.1038/sj.jim.2900731. [DOI] [PubMed] [Google Scholar]
- 60.Smith D.J., Martin V.J.J., Mohn W.W. A cytochrome P450 involved in the metabolism of abietane diterpenoids by Pseudomonas abietaniphila BKME-9. J. Bacteriol. 2004;186:3631–3639. doi: 10.1128/JB.186.11.3631-3639.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith D.J., Park J., Tiedje J.M., Mohn W.W. A large gene cluster in Burkholderia xenovorans encoding abietane diterpenoid catabolism. J. Bacteriol. 2007;189:6195–6204. doi: 10.1128/JB.00179-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith D.J., Patrauchan M.A., Florizone C., Eltis L.D., Mohn W.W. Distinct roles for two CYP226 family cytochromes P450 in abietane diterpenoid catabolism by Burkholderia xenovorans LB400. J. Bacteriol. 2008;190:1575–1583. doi: 10.1128/JB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muttray A.F., Yu Z., Mohn W.W. Population dynamics and metabolic activity of Pseudomonas abietaniphila BKME-9 within pulp mill wastewater microbial communities assayed by competitive PCR and RT-PCR. FEMS Microbiol. Ecol. 2001;38:21–31. doi: 10.1111/j.1574-6941.2001.tb00878.x. [DOI] [Google Scholar]
- 64.Burnes T.A., Blanchette R.A., Farrell R.L. Bacterial biodegradation of extractives and patterns of bordered pit membrane attack in pine wood. Appl. Environ. Microbiol. 2000;66:5201–5205. doi: 10.1128/AEM.66.12.5201-5205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kallioinen A., Vaari A., Rättö M., Konn J., Siika-Aho M., Viikari L. Effects of bacterial treatments on wood extractives. J. Biotechnol. 2003;103:67–76. doi: 10.1016/S0168-1656(03)00051-8. [DOI] [PubMed] [Google Scholar]
- 66.Cámara B., Strömpl C., Verbarg S., Spröer C., Pieper D.H., Tindall B.J. Pseudomonas reinekei sp. nov., Pseudomonas moorei sp. nov. and Pseudomonas mohnii sp. nov., novel species capable of degrading chlorosalicylates or isopimaric acid. Int. J. Syst. Evol. Microbiol. 2007;57:923–931. doi: 10.1099/ijs.0.64703-0. [DOI] [PubMed] [Google Scholar]
- 67.Wilson A.E., Moore E.R., Mohn W.W. Isolation and characterization of isopimaric acid-degrading bacteria from a sequencing batch reactor. Appl. Environ. Microbiol. 1996;62:3146–3151. doi: 10.1128/aem.62.9.3146-3151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mohn W.W. Bacteria obtained from a sequencing batch reactor that are capable of growth on dehydroabietic acid. Appl. Environ. Microbiol. 1995;61:2145–2150. doi: 10.1128/aem.61.6.2145-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Côté R., Otis C. Étude de la biodégradation de l’acide dehydroabietique par Bacillus psychrophilus. Rev. Des Sci. L’eau. 1989;2:313–324. doi: 10.7202/705033ar. [DOI] [Google Scholar]
- 70.Martin V.J.J., Mohn W.W. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J. Bacteriol. 1999;181:2675–2682. doi: 10.1128/jb.181.9.2675-2682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin V.J.J., Mohn W.W. Genetic investigation of the catabolic pathway for degradation of abietane diterpenoids by Pseudomonas abietaniphila BKME-9. J. Bacteriol. 2000;182:3784–3793. doi: 10.1128/JB.182.13.3784-3793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morgan C.A., Wyndham R.C. Characterization of tdt genes for the degradation of tricyclic diterpenes by Pseudomonas diterpeniphila A19-6a. Can. J. Microbiol. 2002;48:49–59. doi: 10.1139/w01-127. [DOI] [PubMed] [Google Scholar]
- 73.Eaton R.W., Chapman P.J. Bacterial metabolism of naphthalene: Construction and use of recombinant bacteria to stody ring cleavage os 1,2-dihydroxynaphthalene and subsequent reactions. J. Bacteriol. 1992;174:7542–7554. doi: 10.1128/jb.174.23.7542-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tavendale M.H., McFarlane P.N., Mackie K.L., Wilkins A.L., Langdon A.G. The fate of resin acids-1. The biotransformation and degradation of deuterium labelled dehydroabietic acid in anaerobic sediments. Chemosphere. 1997;35:2137–2151. doi: 10.1016/S0045-6535(97)00293-2. [DOI] [Google Scholar]
- 75.Tavendale M.H., McFarlane P.N., Mackie K.L., Wilkins A.L., Langdon A.G. The fate of resin acids-2. The fate of resin acids and resin acid derived neutral compounds in anaerobic sediments. Chemosphere. 1997;35:2153–2166. doi: 10.1016/S0045-6535(97)00294-4. [DOI] [Google Scholar]
- 76.Meyer T., Yang M.I., Tran H.N., Allen D.G., Edwards E.A. Impact of resin and fatty acids on full-scale anaerobic treatment of pulp and paper mill effluents. Environ. Eng. Sci. 2016;33:394–403. doi: 10.1089/ees.2015.0395. [DOI] [Google Scholar]
- 77.Aguirre M., Vuorenmaa J., Valkonen E., Kettunen H., Callens C., Haesebrouck F., Ducatelle R., Van Immerseel F., Goossens E. In-feed resin acids reduce matrix metalloproteinase activity in the ileal mucosa of healthy broilers without inducing major effects on the gut microbiota. Vet. Res. 2019;50:1–14. doi: 10.1186/s13567-019-0633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Savluchinske-Feio S., Curto M.J.M., Gigante B., Roseiro J.C. Antimicrobial activity of resin acid derivatives. Appl. Microbiol. Biotechnol. 2006;72:430–436. doi: 10.1007/s00253-006-0517-0. [DOI] [PubMed] [Google Scholar]
- 79.Savluchinske-Feio S., Nunes L., Tavares P., Silva A.M., Roseiro J.C., Gigante B., João M., Curto M. Activity of dehydroabietic acid derivatives against wood contaminant fungi. J. Microbiol. Methods. 2007;70:465–470. doi: 10.1016/j.mimet.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 80.Tolmacheva I.A., Tarantin A.V., Boteva A.A., Anikina L.V., Vikharev Y., Grishko V.V., Tolstikov A.G. Synthesis and biological activity of nitrogen-containing derivatives of methyl dehydroabietate. Pharm. Chem. J. 2006;40:489–493. doi: 10.1007/s11094-006-0161-0. [DOI] [Google Scholar]
- 81.Xu D.D., Yan Y., Jiang C.X., Liang J.J., Li H.F., Wu Q.X., Zhu Y. Sesquiterpenes and diterpenes with cytotoxic activities from the aerial parts of Carpesium humile. Fitoterapia. 2018;128:50–56. doi: 10.1016/j.fitote.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 82.Lee T.K., Park J.Y., Yu J.S., Jang T.S., Oh S.T., Pang C., Ko Y.J., Kang K.S., Kim K.H. 7α,15-Dihydroxydehydroabietic acid from Pinus koraiensis inhibits the promotion of angiogenesis through downregulation of VEGF, p-Akt and p-ERK in HUVECs. Bioorganic Med. Chem. Lett. 2018;28:1084–1089. doi: 10.1016/j.bmcl.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 83.González M. Aromatic abietane diterpenoids: Their biological activity and synthesis. Nat. Prod. Rep. 2015:684–704. doi: 10.1039/C4NP00110A. [DOI] [PubMed] [Google Scholar]
- 84.Parimal K., Khale A., Pramod K. Resins from herbal origin and a focus on their applications. Int. J. Pharm. Sci. Res. 2011;2:1077–1085. [Google Scholar]
- 85.Yadav B.K., Gidwani B., Vyas A. Rosin: Recent advances and potential applications in novel drug delivery system. J. Bioact. Compat. Polym. 2015:1–16. doi: 10.1177/0883911515601867. [DOI] [Google Scholar]
- 86.Gonzalez M.A. Aromatic abietane diterpenoids: Total syntheses and synthetic studies. Tetrahedron. 2015;71:1883–1908. doi: 10.1016/j.tet.2015.01.058. [DOI] [Google Scholar]
- 87.Kugler S., Ossowicz P., Malarczyk-Matusiak K., Wierzbicka E. Advances in rosin-based chemicals: The latest recipes, applications and future trends. Molecules. 2019;24:1651. doi: 10.3390/molecules24091651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loughlin W.A. Biotransformations in organic synthesis. Bioresour. Technol. 2000;74:49–62. doi: 10.1016/S0960-8524(99)00145-5. [DOI] [Google Scholar]
- 89.Huang F.X., Yang W.Z., Ye F., Tian J.Y., Hu H.B., Feng L.M., Guo D.A., Ye M. Microbial transformation of ursolic acid by Syncephalastrum racemosum (Cohn) Schroter AS 3.264. Phytochemistry. 2012;82:56–60. doi: 10.1016/j.phytochem.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 90.Gouiric S.C., Feresin G.E., Tapia A.A., Rossomando P.C., Schmeda-Hirschmann G., Bustos D.A. 1b,7b-Dihydroxydehydroabietic acid, a new biotransformation product of dehydroabietic acid by Aspergillus niger. World J. Microbiol. Biotechol. 2004;20:281–284. doi: 10.1023/B:WIBI.0000023834.60165.79. [DOI] [Google Scholar]
- 91.Özşen Ö., Kıran İ., Dağ İ., Atlı Ö., Çiftçi G.A., Demirci F. Biotransformation of abietic acid by fungi and biological evaluation of its metabolites. Process Biochem. 2017;52:130–140. doi: 10.1016/j.procbio.2016.09.022. [DOI] [Google Scholar]
- 92.Tapia A.A., Vallejo M.D., Gouiric S.C., Feresin G.E., Rossomando P.C., Bustos D.A. Hydroxylation of dehydroabietic acid by Fusarium species. Phytochemistry. 1997;46:131–133. doi: 10.1016/S0031-9422(97)00207-0. [DOI] [PubMed] [Google Scholar]
- 93.Van Beek T.A., Claassen F.W., Dorado J., Godejohann M., Sierra-Alvarez R., Wijnberg J.B.P.A. Fungal biotransformation products of dehydroabietic acid. J. Nat. Prod. 2007;70:154–159. doi: 10.1021/np060325e. [DOI] [PubMed] [Google Scholar]
- 94.Yang N.-Y., Liu L., Tao W.-W., Duan J.-A., Tian L.-J. Phytochemistry diterpenoids from Pinus massoniana resin and their cytotoxicity against A431 and A549 cells. Phytochemistry. 2010;71:1528–1533. doi: 10.1016/j.phytochem.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 95.Witzig R., Aly H.A.H., Strömpl C., Wray V., Junca H., Pieper D.H. Molecular detection and diversity of novel diterpenoid dioxygenase DitA1 genes from proteobacterial strains and soil samples. Environ. Microbiol. 2007;9:1202–1218. doi: 10.1111/j.1462-2920.2007.01242.x. [DOI] [PubMed] [Google Scholar]
- 96.Bleif S., Hannemann F., Lisurek M., von Kries J.P., Zapp J., Dietzen M., Antes I., Bernhardt R. Identification of CYP106A2 as a regioselective allylic bacterial diterpene hydroxylase. ChemBioChem. 2011;12:576–582. doi: 10.1002/cbic.201000404. [DOI] [PubMed] [Google Scholar]
- 97.Janocha S., Zapp J., Hutter M., Kleser M., Bohlmann J., Bernhardt R. Resin acid conversion with CYP105A1: An enzyme with potential for the production of pharmaceutically relevant diterpenoids. ChemBioChem. 2013;14:467–473. doi: 10.1002/cbic.201200729. [DOI] [PubMed] [Google Scholar]