Figure 1.
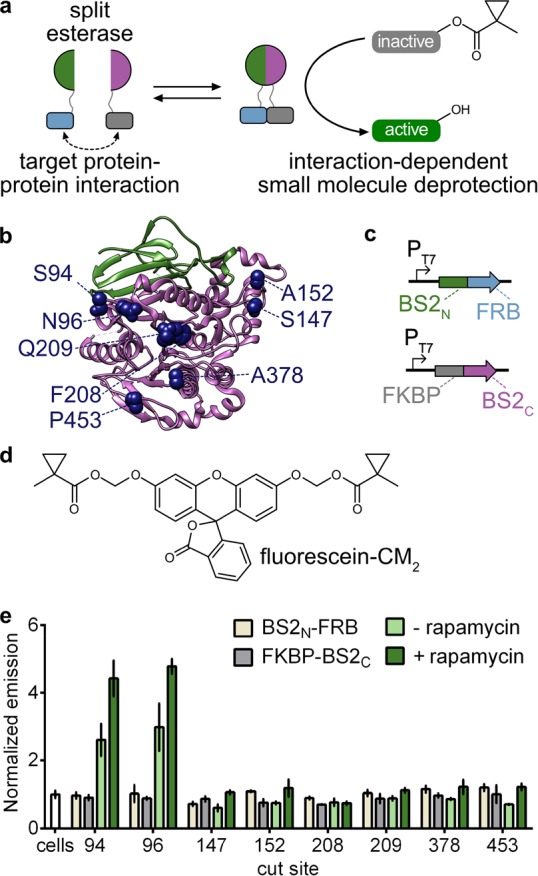
Development of a split esterase sensor. (a) Schematic of a PPI-driven split esterase assembly to unmask 1-methylcyclopropyl (CM)-masked molecules. Interacting proteins are fused to inactive fragments of BS2 esterase. Interaction between the fusion proteins results in assembly of the esterase which cleaves an inactive small molecule to generate an active molecule and output signal. (b) Mapping the cut sites onto a homologous B. subtilis esterase structure (PDB 1QE3). The BS2N fragment (green) and BS2C (magenta) from the lead split site, S94, are shown. Split sites occur after the designated amino acids. (c) Vector system to identify PPI-dependent esterase fragments in E. coli. (d) Chemical structure of masked fluorophore fluorescein-CM2. (e) Fluorescence output of split esterase fragments. E. coli expressing BS2N-fused FRB (tan), FKBP-fused BS2C (gray), or both in the absence (light green) or presence (green) of rapamycin were incubated with fluorescein-CM2 for 4 h and then analyzed for fluorescence. Error bars are the standard deviation for n = 3 replicates.
