Currently, there are more than 216 million annotated protein sequences available in public databases, a number that doubles every 28 months, and just like the deep sea floor, only a minuscule portion of this territory has been explored.1 Each sequence encodes for a protein with a unique composition and order of amino acids that dictate its fold, and in the case of an enzyme, the reactions it can catalyze. However, predicting function based on sequence is not an easy feat. Typically, function has been experimentally determined through labor-intensive protein expression and isolation coupled with experimental characterization of enzymes from primary metabolism and natural product biosynthetic pathways. In this issue of ACS Central Science, Lewis and co-workers survey the activity across one family of enzymes in order to profile reactivity and selectivity across a range of substrates.2
Well-characterized enzymes have historically served as benchmarks for predicting function of uncharacterized enzymes. For example, flavin-dependent monooxygenases (FDMOs) can mediate various transformations depending on their fold. One known function for a subset of monooxygenases, class F flavin adenine dinucleotide (FAD)-dependent monooxygenases, is halogenation.3 This class of enzymes shares a structurally similar nucleotide binding site to class A aromatic hydroxylases; however, a unique tryptophan cage provides class F FAD-dependent monooxygenases with a characteristic sequence fingerprint. To predict function and mechanism, experimental findings on class F FDMOs coupled with the amino acid fingerprint are often applied to related sequences. While this approach can lead to accurate function assignments in some cases, there other instances in which enzymes possess slightly altered motifs and can be overlooked in such a function assignment.
By constructing a sequence similarity network (SSN)4 containing sequences with the highest similarity to well-studied flavin-dependent halogenases (FDHs) involved in indole alkaloid biosynthesis, the authors define the sequence space hypothesized to have conserved halogenase activity. This SSN contained nearly 4000 sequences, of which 129 had been previously characterized. Lewis and co-workers canvassed the FDH family and identified 128 putative FDHs based on a sequence motif conserved across characterized halogenases. To profile how these “unknown” sequences fit within the family, the authors profiled the activity of these enzymes against a panel of substrates and halide sources. This allowed the researchers to identify trends in reactivity across this family of enzymes and ultimately identify wild-type enzymes capable of halogenating previously intractable substrates in a site-selective manner.
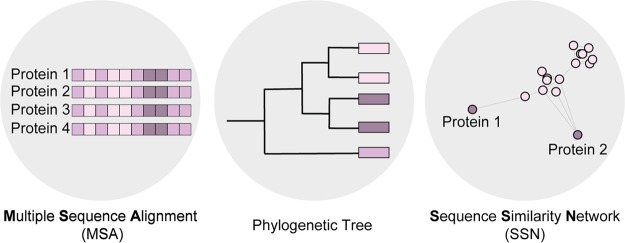
There has been an evolution of the tools available for canvassing and identifying sequence space with untapped synthetic potential. Some of the commonly used tools for sequence profiling and visualization are multiple sequence alignment,5 phylogenetic trees,6 and sequence similarity networks7 (Figure 1). Additionally, there are other visualization tools being developed such as the variational autoencoder latent space model.8
Figure 1.
Tools for sequence profiling from left to right: multiple sequence alignment, phylogenetic tree, and sequence similarity network.
In a multiple sequence alignment, three or more protein sequences that have some evolutionary connection are aligned (Figure 1). This profiling can be used to identify functional relationships among sequences. This approach highlights conserved motifs that can potentially be used to predict enzyme function or pinpoint residues that might be important for enzyme function
Phylogenetic trees, as previously mentioned, indicate the relationship between sequences across evolution (Figure 1). The branching pattern of these trees reflects how proteins evolved from a series of common ancestors. This tool can illuminate which sequences within a family are most related and distinguish close cousins from distant relatives. However, performing family-wide profiling requires an accurate large-scale sequence alignment, which can be challenging.
Visualizing sequences relationships for family -wide profiling can be cumbersome with the methods previously outlined. Sequence similarity networks are visual tools that were developed to group protein sequences based on a similarity threshold. Depending on this threshold for similarity, the sequences can be grouped based on their homology, which can translate to their potential reactivity. For example, the original SSN constructed by Lewis and co-workers revealed a clustering of sequences based on their native substrate, with FDHs known to halogenate phenols and FDHs that naturally modify tryptophan substrates, each forming separate groupings.
In this study, 128 putative sequences from across the FDH sequence space defined by the SSN were obtained as codon-optimized genes. From this set, 87 proteins were successfully expressed in yields sufficient for reactivity screening. By testing the reactivity of this enzyme panel with 12 substrates, the authors began to fill in the vast reactivity gaps across this enzyme family and establish reactivity leads that could be exploited through further profiling of the related sequence space or established protein engineering methods.
A recent study by Goss and co-workers further highlights the synthetic benefit of FDH family-wide profiling.9 They reported the first FDH that iodinates in vitro identified through family-wide profiling using a previously unappreciated sequence motif. This serves as a great example of the potential of underexplored FDHs regions, which merits further investigation. In other studies, SSNs have proven useful for profiling other classes of FAD-dependent enzymes to identify biocatalysts appropriate for target-oriented synthesis.10
These examples showcase the synthetic utility of enzymes hiding in plain sight—the sequences are known, but their reactivity will remain a mystery without dedicated experimental work toward family-wide reactivity profiling. These efforts are guided by tools for visualizing sequence space and have the potential to bring light to the deep sea floor of unexplored enzymes.
The authors declare no competing financial interest.
References
- GenBank and WGS Statistics. https://www.ncbi.nlm.nih.gov/genbank/statistics/ (accessed October 30, 2019).
- Fisher B. F.; Snodgrass H. M.; Jones K. A.; Andorfer M. C.; Lewis J. C. Site-Selective C–H Halogenation Using Flavin-Dependent Halogenases Identified via Family-Wide Activity Profiling. ACS Cent. Sci. 2019, 10.1021/acscentsci.9b00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers M. M. E.; Montersino S.; Westphal A. H.; Tischler D.; Van Berkel W. J. H. Flavin Dependent Monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. 10.1016/j.abb.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Gerlt J. A.; Bouvier J. T.; Davidson D. B.; Imker H. J.; Sadkhin B.; Slater D. R.; Whalen K. L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A Web Tool for Generating Protein Sequence Similarity Networks. Biochim. Biophys. Acta 2015, 1854 (8), 1019–1037. 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F.; Park Y. mi; Lee J.; Buso N.; Gur T.; Madhusoodanan N.; Basutkar P.; Tivey A. R. N.; Potter S. C.; Finn R. D.; Lopez R. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47 (W1), W636–W641. 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos G. A.; Soldatos T. G.; Barbosa-Silva A.; Schneider R.. A Reference Guide for Tree Analysis and Visualization. BioData Min. 2010, 3( (1), ), 10.1186/1756-0381-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R.; Oberg N.; Gerlt J. A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58 (41), 4169–4182. 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.. Ph.D. Thesis, University of Michigan, 2019, https://deepblue.lib.umich.edu/handle/2027.42/147634.
- Gkotsi D. S.; Ludewig H.; Sharma S. V.; Connolly J. A.; Dhaliwal J.; Wang Y.; Unsworth W. P.; Taylor R. J. K.; McLachlan M. M. W.; Shanahan S.; Naismith J. H.; Goss R. J. M.. A Marine Viral Halogenase That Iodinates Diverse Substrates. Nat. Chem. 2019, 10.1038/s41557-019-0349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyser J. B.; Baker Dockrey S. A.; Benitez A. R.; Joyce L. A.; Wiscons R. A.; Smith J. L.; Narayan A. R. H.. Stereodivergent, Chemoenzymatic Synthesis of Azaphilone Natural Products. J. Am. Chem. Soc. 2019, 141, 10.1021/jacs.9b09385. [DOI] [PMC free article] [PubMed] [Google Scholar]