Figure 1.
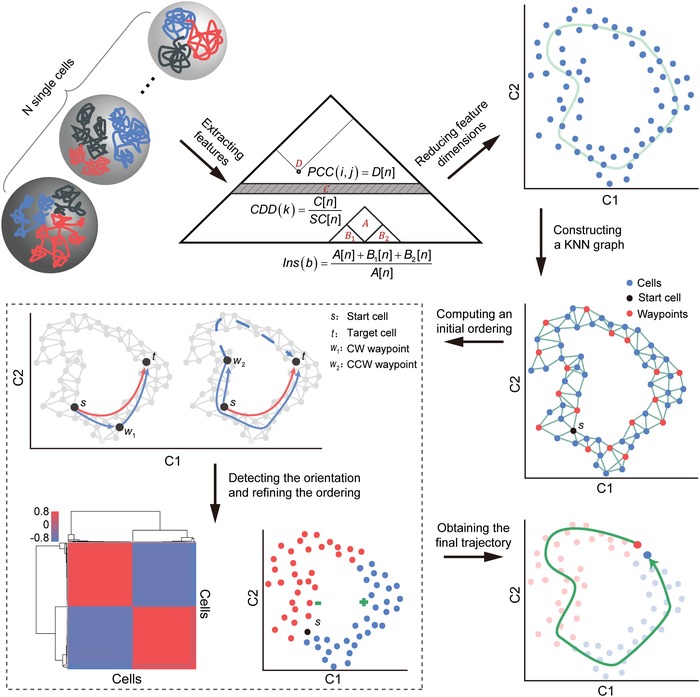
Illustration of CIRCLET for reconstructing a cell‐cycle trajectory from single‐cell Hi‐C maps. CIRCLET contains six key steps. 1) Extracting features: multiscale feature sets are extracted from single‐cell Hi‐C maps. 2) Reducing feature dimensions: the dimension of these feature sets are further reduced to a low n‐dimensional space via diffusion maps (e.g., 2D as an example). 3) Constructing a KNN graph: CIRCLET constructs a k‐nearest‐neighbor graph in the n‐dimensional embedded space and selects a set of cells called “waypoints,” one of which is randomly selected as the starting cell s. 4) Computing an initial ordering: an initial ordering of cells is obtained by the shortest path distance from s (e.g., distance D s,t marked by a red solid line from s to t cell). 5) Detecting the orientation and refining the ordering: CIRCLET also computes a perspective matrix P, which records the shortest path distance of each cell to the starting cell s from the viewpoint of waypoints (e.g., the distance of cell t to s from the viewpoint of w 1 is ). These waypoints' perspective is first used to identify the clockwise (CW) or counterclockwise (CCW) semicircle of cells from s. 6) Obtaining the final trajectory: CIRCLET iteratively executes step (5) until convergence, eventually obtaining a high‐resolution cell‐cycle trajectory.
