Figure 2.
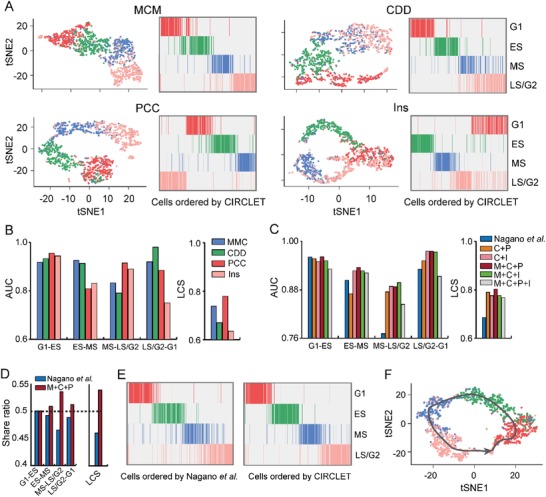
Exploring the preference of multiple feature sets and their combinations. A) tSNE maps and cell heatmaps of four FACS‐sorted cell phases (G1, ES, MS, and LS/G2) ordered based on the reconstructed trajectory by CIRCLET based on four individual feature sets including MCM (M), CDD (C), PCC (P), and Ins (I), respectively. B) Comparison of five evaluation indexes for the reconstructed trajectory by CIRCLET based on the four individual feature sets, respectively. These evaluation indexes include AUC scores between two successive cell‐cycle phases (denoted as G1–ES, ES–MS, MS–LS/G2, and LS/G2–G1) and LCS for measuring labels inconsistency of adjacent cells on the entire inferred trajectory. C) Comparison of the five evaluation indexes for the reconstructed trajectory by CIRCLET by Nagano et al. and five combinations of multiple feature sets. D) The share ratio of five scores between the inferred trajectories by Nagano et al. and CIRCLET based on the best combination of feature sets. E) The reconstructed cell‐cycle trajectories by Nagano et al. (left panel) and CIRCLET (right panel), respectively, based on the best combination (MCM+CDD+PCC) from four FACS‐sorted cells (G1, ES, MS, and LS/G2). F) tSNE maps by CIRCLET based on the best combination (MCM+CDD+PCC) from four FACS‐sorted cell phases (G1, ES, MS, and LS/G2).
