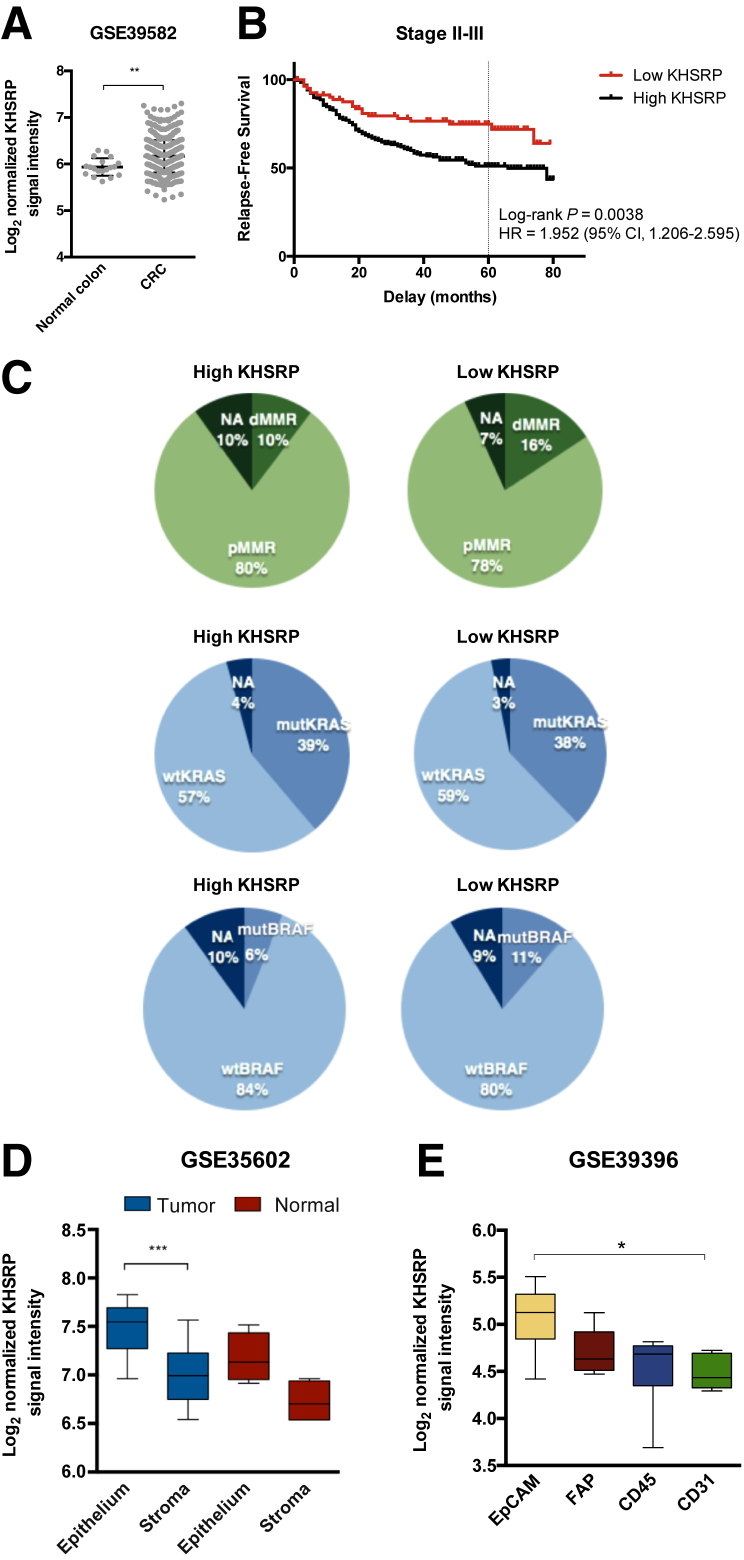
Figure 3.
In silico analysis of three independent cohorts validates the overexpression and prognostic significance of KHSRP. A: Log2 normalized signal intensity of KHSRP in a cohort of 566 colorectal cancer (CRC) patients and 19 normal controls, profiled by gene expression microarrays. Individual points are shown. A Mann-Whitney test was performed. B: Survival curves for patients segregated into high/low groups using the 25th percentile of the KHSRP probe signal intensity. The hazard ratio (HR) and associated CI for relapse-free survival at 5 years (dashed line) are shown. C: Distribution of patients with high/low KHSRP expression, according to clinical characteristics: mismatch repair deficient (dMMR; ie, microsatellite instable) or proficient (pMMR; ie, microsatellite stable), KRAS (codons 12 and 13) mutational status, and BRAF (p.V600E) mutational status. D: Expression of KHSRP in laser-capture microdissected epithelial and stromal cells from 13 CRC and normal colonic mucosa samples. A Kruskal-Wallis test was performed. E: Expression of KHSRP in a cohort of fluorescence-activated cell sorted cells from 14 CRC patients. CD45 indicates leukocytes, and CD31 indicates endothelial cells. A Kruskal-Wallis test was performed. D and E: The box-and-whisker plots depict interquartile range, median, minimum, and maximum limits. Data are expressed as means ± SD (A). *P < 0.05, **P < 0.01, and ***P < 0.01. EpCAM, epithelial cancer cell; FAP, cancer-associated fibroblast; mut-, mutant; NA, not available; wt-, wild-types.