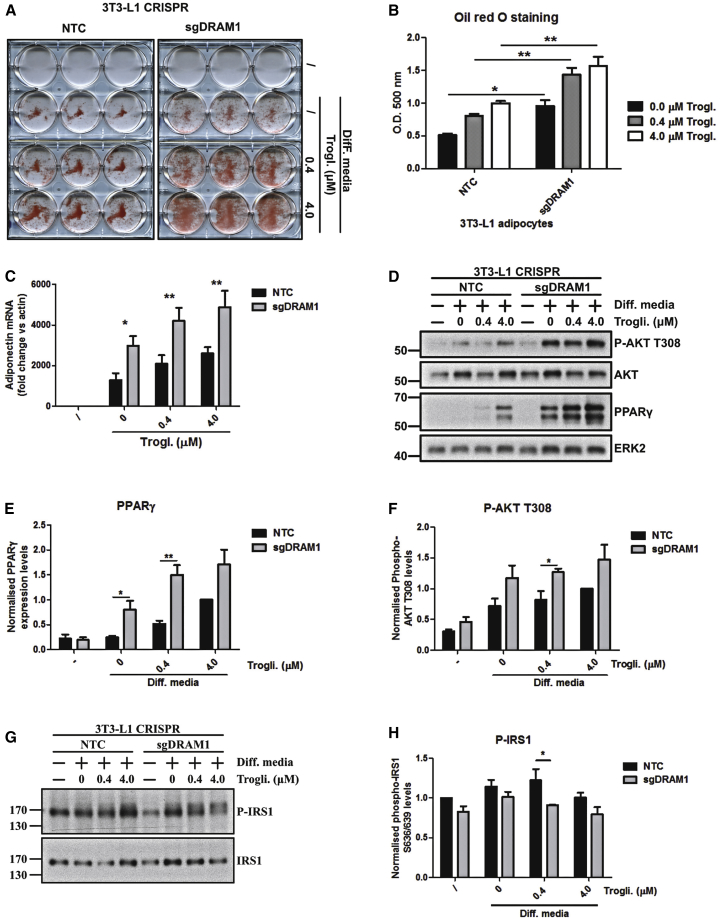
Figure 7.
DRAM1 Impairs 3T3-L1 Pre-adipocyte Differentiation
3T3-L1 pre-adipocytes infected with 2 different guide sequences targeting Dram1 or 2 non-targeting control guides (NTC) were left undifferentiated or differentiated for 8 days in differentiation media containing various amounts of troglitazone (0–4 μM).
(A) Cellular neutral lipid contents were shown following oil red O staining.
(B) Staining was quantified by eluting oil red O in isopropanol and measuring optical densities at a wavelength of 500 nm. Results are the mean staining of the 2 different Dram1 CRISPR cell lines for each condition normalized to the average staining of the 2 different NTC cell lines. Results are from 3 independent experiments.
(C) qPCR analysis showing Adiponectin expression levels in 3T3-L1 NTC or Dram1 CRISPR cells grown with regular or differentiation medium containing the indicated concentration of troglitazone.
(D) Pre-adipocyte differentiation efficiency was evaluated by western blots directed against phospho-AKT T308 and PPARɣ. AKT and ERK2 were used as loading controls.
(E) Quantification of PPARɣ levels measured from 3 independent experiments was performed using ImageJ.
(F) Quantification of phospho-AKT levels measured from 3 independent experiments was performed using ImageJ.
(G) mTOR activation was evaluated by western blotting for phospho-S636/639 IRS1 in Dram1 CRISPR 3T3-L1 cells or NTC CRISPR cells.
(H) Quantification of phospho-S636/639 IRS1 levels measured from 3 independent experiments was performed using ImageJ.
(B, C, E, F, and H) Data are mean ± SEM. ∗p < 0.05; ∗∗p < 0.01.
See also Figure S7.