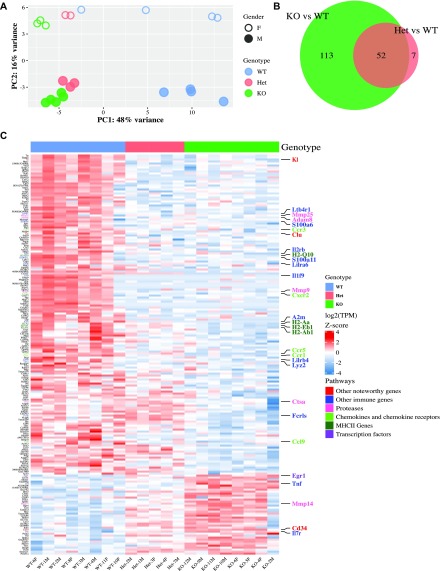
Figure 1. Cx3cr1 loss alters the transcriptome of microglia from 2-mo-old mice.
Microglia were isolated from 2-mo-old Cx3cr1+/+ (WT, n = 8), Cx3cr1+/eGFP (Het, n = 5), or Cx3cr1eGFP/eGFP (KO, n = 8) mice. The RNA isolated from microglia of each mouse was used to generate an individual RNA-seq data set. (A) The PCA indicates that the samples separate by genotype in the first principal component and gender in the second. Each dot represents microglia from an individual mouse. (B) Most of the genes differentially expressed between Het and WT microglia are also differentially expressed by KO and WT microglia. (C) Unsupervised cluster analysis of the top 200 DEGs in microglia from 2-mo-old WT (n = 8), Het (n = 5), or KO (n = 8) mice. Most DEGs are expressed at lower levels in Het and KO samples compared to WT samples. Selected genes and the pathways they are associated with (manual annotation) are represented in different colors.