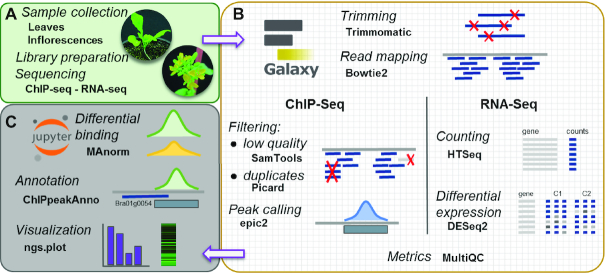
Figure 1:
Schematic view of the analytical workflow of the Reproducible Epigenomic Analysis pipeline (REA). (A) Samples from leaves and inflorescences were used for ChIP-seq and RNA-seq. (B) Major analytical steps were conducted in a reproducible Galaxy workflow, running on a Docker container. (C) Further analysis and graphical representation of results were tracked and run on Jupyter interactive notebooks.