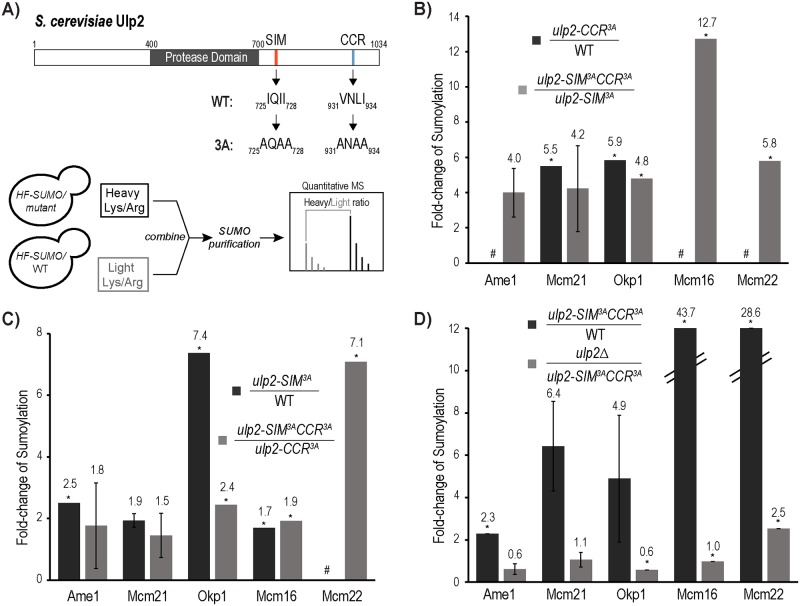
Fig 1. The C-terminal Conserved Region (CCR) and SUMO-interacting motif (SIM) of Ulp2 are both needed to desumoylate the CCAN complex.
A) Illustration depicting Ulp2’s domain structure and the quantitative MS approach used to determine the effect of various ulp2 mutations on sumoylated CCAN in the HF-SMT3 (6×His-3×FLAG-Smt3) strain background (see experimental methods for details). Conserved hydrophobic residues in Ulp2’s SIM and CCR are indicated. Triple alanine mutations of these residues generate the ulp2-SIM3A and ulp2-CCR3A mutants. B-D) Effects of various ulp2 mutations on sumoylated CCAN subunits are shown, while the rest of the MS results can be found in S1–S6 Tables. In each case, the fold-changes in the sumoylated CCAN subunit are shown for each of the indicated strains. CCAN subunits not identified in the MS experiments are indicated by #. Asterisks (*) indicate where an insufficient number of peptides are available for statistical analysis.