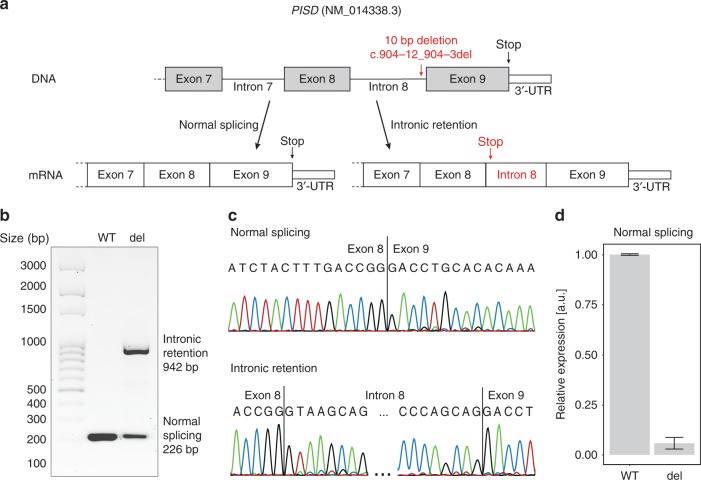
Fig. 3.
Schematic view of thePISDgene with the position of the variant identified and the splicing alteration resulting from it. (a) The variant is a 10-bp deletion in intron 8, close to the splice acceptor site of the last exon. The change in the splicing consensus sequence at this site leads to the production of both a canonical transcript and an aberrant messenger RNA (mRNA) retaining intron 8, which in turn leads to a premature termination of the open reading frame. (b) Electrophoresis of the reverse transcription polymerase chain reaction (RT-PCR) products from HEK293T transfected with a minigene-bearing plasmid, carrying exons 8 to 9 of PISD and the 10-bp deletion (del) or the wild-type sequence (WT). The mutant form of the plasmid reveals two transcripts, corresponding to the normally spliced isoform (226 bp) and the one carrying the intronic retention (942 bp) depicted above. (c) Sanger sequencing of the two transcripts, from the WT minigene and the one carrying the deletion, respectively. (d) Quantification of spliced transcripts originating from the WT or mutant plasmid (del), by quantitative PCR (qPCR). The difference between these two values is statistically significant (p-value = 2.5 × 10−4, by t-test). Error bars represent standard deviation values.