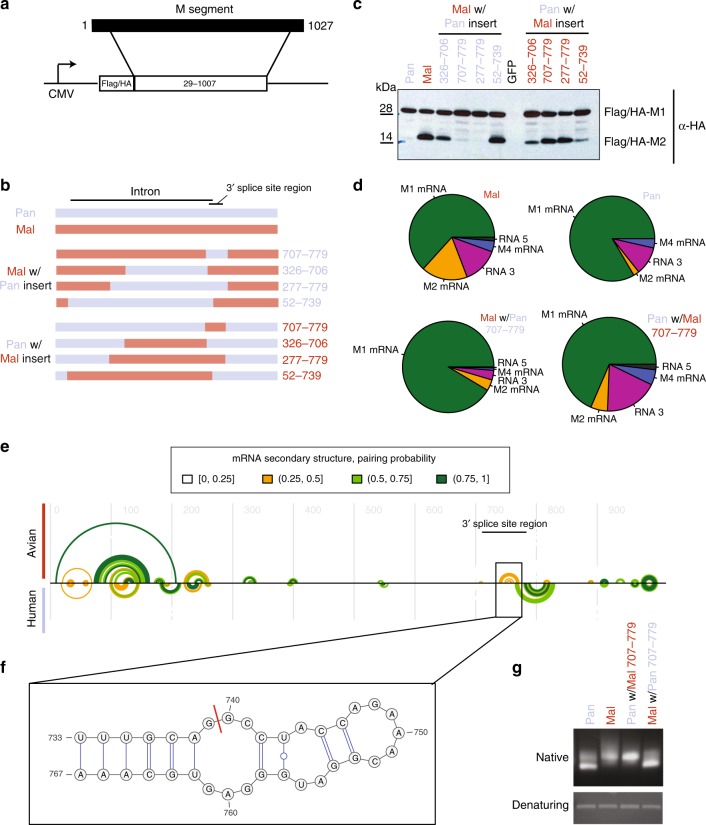
Fig. 5. A cis-regulatory element in the 3′ splice site region controls M segment splicing.
a Reporter system. The coding sequence of segment 7 was cloned into a eukaryotic expression vector with N-terminal Flag/HA tag. b Wild-type Pan and Mal sequences as well as several chimeric Pan/Mal constructs were cloned into the reporter vector. c A549 cells were transfected with the indicated reporter constructs, harvested, and subjected to anti-HA immunoblotting. d HEK293T cells were transfected with constructs encoding NP, PA, PB1, and PB2 (of strain WSN) and vectors encoding for wild-type or chimeric M segments. At 24 h post transfection, RNA was isolated and subjected to RNA-seq. The pies depict the relative counts of isoforms as quantified by splice junction reads normalized to the levels of NP and polymerase subunits. The area of the pies scales with the total number of normalized splice junction reads. The average of n = 2 biological replicates is depicted. e Evolutionary conserved RNA secondary structure along the M mRNA predicted for avian-adapted (top) and human-adapted H3N2 (bottom) IAV strains. Each semi-circle corresponds to 1 bp involving the corresponding two alignment positions. Color coding of base pairs according to their corresponding, estimated base-pairing probability. The 3′ splice site region (nt 707–779) is annotated with a bar. f The insert shows a detail of the predicted RNA secondary structure at the 3′ splice site, predicted for avian but not human sequences in this study. The splice position is indicated by a red bar. g The region predicted to be structured with the two species-specific and mutually exclusive elements (707–825) was in vitro transcribed. The RNA was purified and run on native and denaturing agarose gels stained with ethidium bromide.