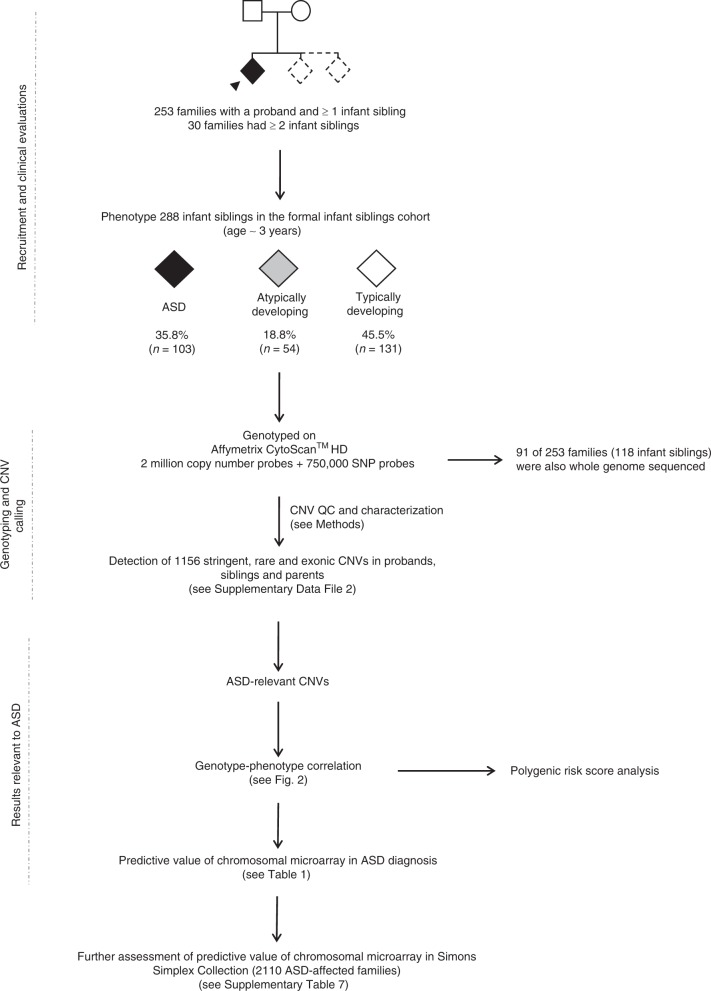
Fig. 1.
Project flowchart. Families consisting of a proband and at least 1 infant sibling were recruited through the Baby Siblings Research Consortium. The proband was the first in the family to receive an ASD diagnosis. Psychometric data were collected for all siblings at ~36 months of age, at which point an ASD diagnosis was made if the child met clinical criteria (see Methods). All children were also assessed for ASD, cognitive and adaptive behavioral functioning at least once prior to the 36-month time point. We genotyped individuals from 253 families on the Affymetrix CytoScanTM HD Array and whole-genome sequenced 91 of these families using established pipelines12–14. Copy number variants determined to be ASD-relevant were confirmed with secondary methods. We scrutinized the phenotypes of high-risk infant siblings carrying these ASD-relevant CNVs to determine whether they (i) had ASD, (ii) were atypically developing, or (iii) were neurotypical/non-ASD. There were also 34 siblings who did not meet criteria for formal enrollment but for whom phenotype and microarray data were available. Following the general BSRC strategy, a separate CNV analysis was conducted on 2124 probands and 2423 non-ASD sibs from 2110 ASD-affected families part of the Simons Simplex Collection. Fourteen probands were monozygotic twins. Of the unaffected sibs, 288 were atypically developing. This analysis was performed to further assess the predictive value of chromosomal microarray. ASD autism spectrum disorder, CNV copy number variation.