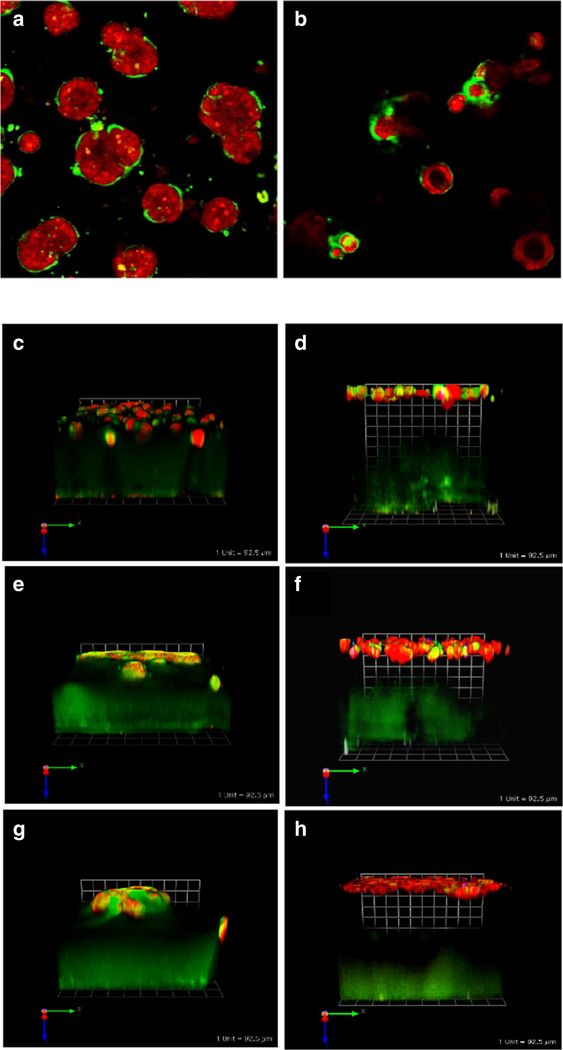
Fig. 3.
Cell/cell interactions in pathomimetic avatars. SUM 225 DCIS cells [42] grow into disorganized dysplastic structures without a central lumen (a). However, in co-cultures with N1ME myoepithelial cells (2 DCIS:1 myoepithelial cell), the structures formed are more organized with some central lumens (b) (cf. with 10A acini in Fig. 2). Cultures were imaged live at 12 days. Two hours before imaging cells were stained with CellTracker Orange (red) to visualize morphology of the structures. Green fluorescence represents degradation products of DQ-collagen IV. Confocal image stacks, obtained on a Zeiss LSM-510 confocal microscope, were rendered in 3D with Volocity software with the equatorial plane shown here; magnification, × 40. Myoepithelial cells also reduce CAF-induced increases in invasiveness and associated degradation of extracellular matrices by MCF-10A, MCF10.DCIS, and MCF10.CA1d grown in tripartite 3D co-cultures. Tripartite cultures consist of CAFs (WS12Ti) plated in collagen I spiked with DQ-collagen I, covered with rBM spiked with DQ-collagen IV that contains cells of MCF10 variants ± myoepithelial (N1ME) cells, and followed by an rBM overlay: c 10A + CAFs, d 10A + CAFs + myoepithelial cells, e 10.DCIS + CAFs, f 10.DCIS + CAFs + myoepithelial cells, g CA1d+ CAFs, and h CA1d + CAFs + myoepithelial cells. Co-cultures were imaged live at day 8. Two hours before imaging cells were stained with CellTracker Orange (red) to visualize morphology of the structures. Green fluorescence represents degradation products of DQ-collagens. Confocal image stacks, obtained on a Zeiss LSM-510 confocal microscope, were rendered in 3D with Volocity software; one grid is 92 μm