Abstract
Glycans and glycoconjugates feature on the “front line” of bacterial cells, playing critical roles in the mechanical and chemical stability of the microorganisms, and orchestrating interactions with the environment and all other living organisms. To negotiate such central tasks, bacterial glycomes incorporate a dizzying array of carbohydrate building blocks and non-carbohydrate modifications, which create opportunities for infinite structural variation. This review highlights some of the challenges and opportunities for the chemical biology community in the field of bacterial glycobiology.
Keywords: bacterial glycome; carbohydrate database; metabolic engineering; glycan array; heptose-1,7-bisphosphate; antibiotic resistance
Introduction
The human glycome is frequently deemed to be a labyrinthine nightmare. Those glycans are based on 10 unique monosaccharide building blocks, which can be combined into oligomers and polymers with linear and branched structures joined by glycosidic linkages between the various hydroxyl groups, with each glycosidic linkage adopting either α- or β-anomeric configuration. The structures may additionally be subject to modifications including, for example, sulfation and phosphorylation [1,2]. Despite this complexity however, when it comes to comparisons of carbohydrate diversity between prokaryotic and eukaryotic life forms, prokaryotes win hands-down. There are hundreds of unique monosaccharide building blocks and chemically-defined saccharide modifications in the database of naturally-occurring carbohydrates (http://csdb.glycoscience.ru/bacterial/) [3,4]; the vast majority of these are prokaryote-specific. The carbohydrates that feature in bacterial glycans range from trioses to dodecoses (Figure 1a) and amongst these, the hexoses show extensive variation in substitution patterns and the heptoses and octoses are exclusively prokaryotic. The nonulosonic acids, which are derived biosynthetically from the hexoses, are also far more varied: there is a single example in man, but dozens have been characterized to date in various bacteria [5]. In addition, a plethora of modifications including alkyl, acyl, amino acyl, phosphoryl and even nucleoside groups are often found decorating the carbohydrates [4]. Defining and cataloging this variety, not to mention understanding its significance, is a Herculean task.
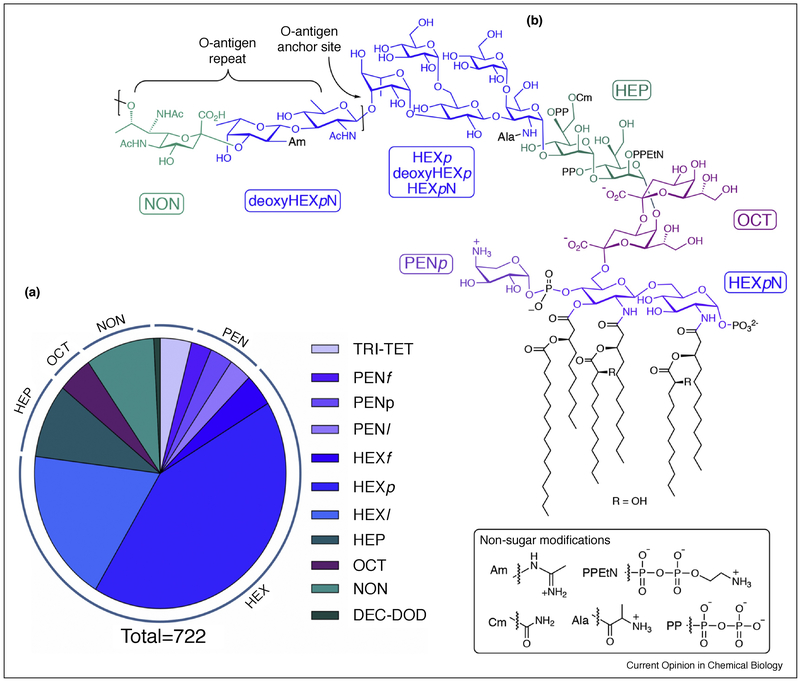
Figure 1.
Bacterial carbohydrate diversity.
(a) Bacterial carbohydrate distribution based on size. The current BCSDB (http://csdb.glycoscience.ru/bacterial/) includes over 700 unique carbohydrates ranging from trioses to dodecanoses [4]. Pentoses and hexoses are further differentiated as furanose (f), pyranose (p) and acyclic/unknown (l). (b) The structure of a lipopolysaccharide (LPS smooth) from Pseudomonas aeruginosa (O12 serotype) exemplifies the diversity of carbohydrates in a single glycoconjugate [6]. O-Acetyl groups are not shown. An Arap4NP is shown on the Lipid A diglucosamine core for the purpose of illustrating the modification. This carbohydrate is known to be associated with P. aeruginosa, but not specifically documented in the serotype illustrated. Further glycan variation is achieved by carbohydrate modifications for example with amidino (Am), carbamoyl (Ca), phosphatidyl ethanolamine (PEtN), diphosphate (PP) and alanyl (Ala).
Bacterial carbohydrates may be components of repeating glycopolymers or, more complex glycoconjugates, which may reveal elaborate “samplers” of different sugars (Figure 1b) [6]. In cells, the glycans commonly include cell-surface structures that are essential for the mechanical integrity of bacterial cells and for critical interactions with other bacteria and the hosts with which they coexist [7,8]. The key glycoconjugates in Gram-negative bacteria are peptidoglycan (PG), lipopolysaccharide (LPS), lipooligosaccharide (LOS), capsular extracellular polysaccharide (EPS), capsular polysaccharide (CPS), and N- and O-linked glycoproteins. In contrast, Gram-positive organisms lack LPS and LOS but instead feature glycosylated lipo- and wall techoic acids (LTAs and WTAs), which are anionic copolymers of glycerol or ribitol phosphate and carbohydrates linked via phosphodiester linkages, conjugated to either PG (for WTA) or diacyl glycerol (LTA). The Gram-positive mycobacteria show additional glycan variations including arabinogalactan and lipoarabinomannan conjugates, which feature many furanose sugars [9].
This opinion summarizes recent examples of the importance of chemical biology approaches for identifying the fates of bacterial sugars in pathogen glycoconjugates, investigating carbohydrate-based interactions, defining the activity of bacterial monosaccharides in eukaryotic signaling, and highlighting the significance of glycan modifications in antibiotic resistance.
Enlisting metabolic labeling for tracking and analyzing bacterial cell-surface glycomes
Metabolic oligosaccharide engineering (MOE), involves delivery of cell-permeable biosynthetic precursors of glycan constituents into living cells and organisms [10]. MOE reagents are commonly modified with azides or terminal alkynes, which serve as biorthogonal reactivity handles for the introduction of epitope, biotin, or fluorescent tags. MOE has proven extremely valuable in research on the glycobiology of mammalian cells and model organisms. For example, studies with peracetylated derivatives of C-2 acetamido hexoses, in which the N-acetyl groups are replaced with N-azidoacetyl are useful for labeling eukaryotic N- and O-linked glycoproteins [10]. For these efforts, knowledge of cellular pathways for conversion of carbohydrate analogs into nucleotide-activated sugars for glycoconjugate assembly is crucial. Notably, the in-cell conversion of free N-acetyl mannosamine into CMP-N-acetylneuraminic acid (CMP-NeuNAc), a non-2-ulosonic acid (NulO) [11,12] has been extensively exploited as it is commonly associated with mammalian cell-surface glycoproteins. NeuNAc is the only nine-carbon sugar in man with the main variation in non-human mammals being the N-glycolyl derivative [13]. MOE in mammalian cells is also enabled by a salvage pathway that affords phosphorylation of unmodified hexoses to generate precursors for conversion into UDP-sugars [14]. This pathway may not be broadly present bacteria and so cannot always be relied upon for MOE.
Information on the pathways for glycoconjugate biosynthesis in bacteria has led to important avenues for selective MOE in bacterial pathogens. Prokaryote-specific NulOs, feature prominently in cell surface-glycoconjugates. NulO biosynthesis follows a common logic across domains of life, with the key step involving a three-carbon extension of a six-carbon nucleotide sugar with phosphoenol pyruvate (PEP). For example, CMP-pseudaminic acid (CMP-Pse) [15] and CMP-legionaminic acid (CMP-Leg) [16] are biosynthesized from UDP-GlcNAc and GDP-GlcNAc, respectively, via the intermediacy of tri-deoxy-diacetamido-hexoses (Figure 2a). Biochemical manipulation of bacterial hexoses, such as those in the Pse and Leg pathways, commonly involve site-selective oxidation of hydroxyl groups (at C3 or C4), which in turn enables further biochemical manipulation such as elimination, enolization and reprotonation for epimerization or ketoisomerization, and transamination [17].
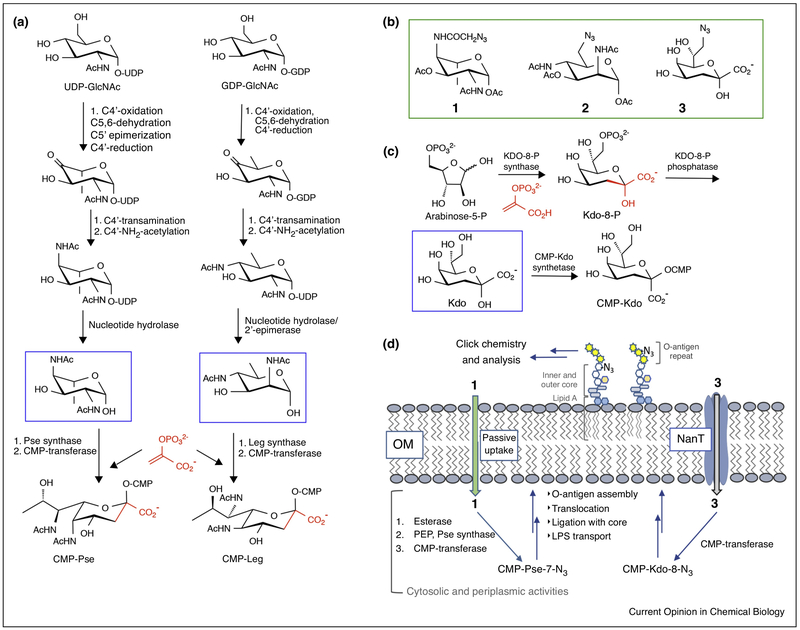
Figure 2.
Knowledge of carbohydrate biosynthetic pathways in bacteria provides important information on precursors for metabolic oligosaccharide engineering (MOE).
(a) Pseudaminic and legionaminic acid biosynthesis via three-carbon extension (PEP – shown in red) of hexose precursors via the Pse (Helicobacter pylori) and Leg (Campylobacter jejuni) pathways. (b) Azide-labeled MOE precursors 1, 2, and 3 for Pse, Leg and Kdo pathways (green box). (c) Octulosonic acid biosynthesis (E. coli) via a three-carbon extension (PEP – shown in red) of a pentose precursor. (d) Cellular uptake and fate of 1 and 3 in bacterial MOE studies.
In bacteria, NulOs play important roles in virulence and pathogenicity and may function in carbohydrate mimicry foiling the human immune system [5]. Bacterial NulOs are common modifications of the capsular polysaccharide (CPS) and O-antigen, and are attached to the Fla proteins in flagellar appendages that are essential for motility. Critical to successful MOE is the provision of cell-permeable precursors, commonly as neutral, per-O/N-acetylated derivatives of hexose or hexosamine intermediates, which are de-O-acetylated by cellular esterases and enter into endogenous pathways for conversion into nucleotide-activated sugars for glycan assembly. Recent MOE studies targeted at the Pse pathway, employing 2- and 4-azidoacetyl analogs of the 2,4-diNAc-L-altrose (2,4-diNAcAlt) intermediate [18], showed superior incorporation of 2-NAc-4NAz-Alt (1, Figure 2b) into LPS and glycoproteins by MOE. Strain-specific metabolic labeling and fluorescence-based imaging of the PA1244 strain of Pseudomonas aeruginosa was also realized. Additionally, earlier MOE on the Leg pathway showed that C-6 azide modification of the neutral 6-deoxyhexose intermediate en route to legionaminic acid (2, Figure 2b), could also be applied for strain-selective labeling of strains of Legionella pneumophilia [19].
MOE has also been applied to 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo), which is found in the inner core of the LPS of Gram-negative bacteria. Kdo is biosynthesized through a PEP-based extension of a pentose sugar (Figure 2c) and incorporation of the 8-azido analog of Kdo (Kdo-8-N3) (3, Figure 2a) into the E. coli LPS has been validated [20]. Recognizing that Kdo is a specialized carbohydrate in Gram-negative bacteria, Chen and coworkers employed MOE with Kdo-8-N3 to distinguish between Gram-negative and Gram-positive bacteria in the mouse gut microbiota [21]. In these studies, Gram-negative bacteria were labeled in vivo by MOE with Kdo-8-N3, followed treatment of fixed tissue with an alkyne-TAMRA reagent for click chemistry (Figure 2d). Gram-positive bacteria were visualized with a fluorescent derivative of vancomycin (Vanco-Bodipy), which binds more readily to the surface-exposed peptidoglycan relative to the periplasmic peptidoglycan of Gram-negative bacteria. Recently, it has been reported that transport of Kdo-8-N3 into E. coli is mediated by the sialic acid transporter NanT and that uptake of Kdo-8-N3 into pathogens, including P. aeruginosa and Acinetobacter baumannii, which lack NanT, can be improved by heterologous expression of E. coli NanT [22]. This genetic manipulation will expand the scope of MOE with Kdo for the study of Gram-negative pathogens.
In summary, the growing understanding bacterial carbohydrate diversity across bacterial phyla suggests that MOE holds considerable promise for the species and strain-selective labeling of bacteria that reaches far beyond the simple crystal violet Gram-stain introduced at the end of the 19th century.
Cellular signaling by bacterial monosaccharides
Recently, there have been intriguing observations on the pathogen-associated molecular pattern (PAMP) activity of soluble bacterial carbohydrates in eukaryotic cells. Host recognition of PAMPs is important for initiating innate immune responses that are critical for eliminating pathogens and for the deployment of adaptive immunity processes [23]. PAMPs are commonly associated with Gram-negative LPS and Gram-positive LTA glycoconjugates.
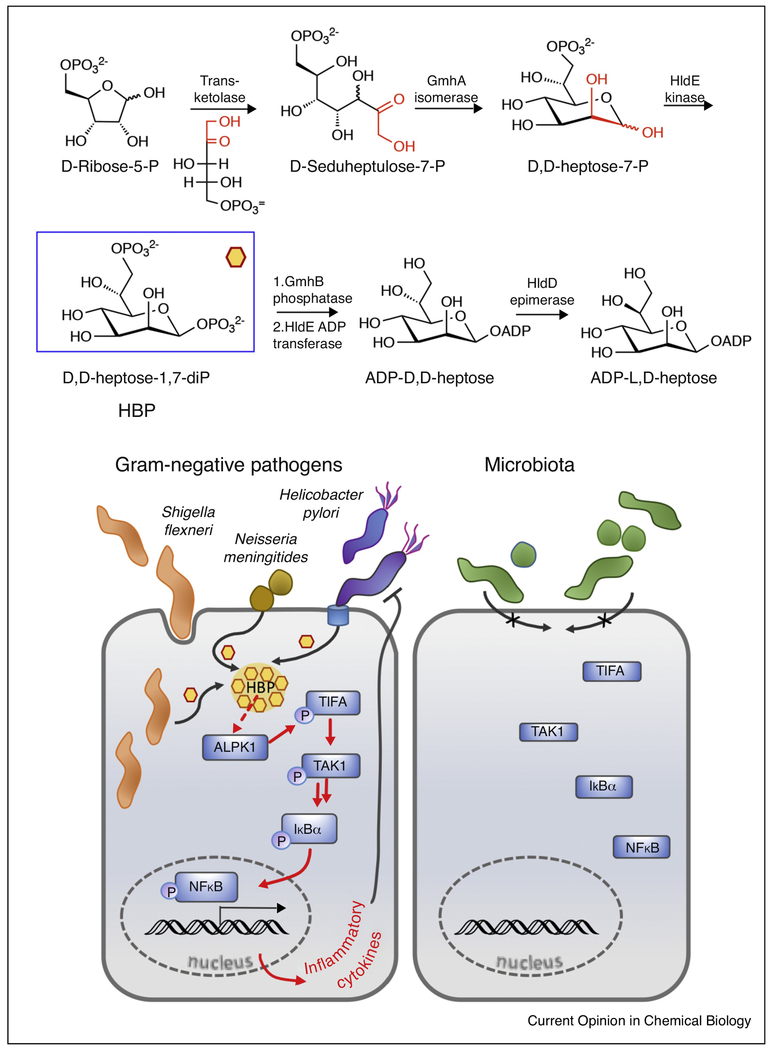
In 2015 it was reported that mammalian cells detect and respond to a heat-resistant factor behaving as a PAMPs [24]. This factor was determined to be heptose-1,7-bisphosphate (HBP). HBP is an intermediate in the biosynthesis of an ADP-heptose, which is a key precursor in the LPS pathway [25] (Figure 3a). Studies in various pathogens revealed that HBP can be delivered into host cells by different mechanisms including via the cag Type 4 secretion system (cag TSS4) of Helicobacter pylori [24,26], by transfer across membranes from Neisseria [24], and by direct secretion from the intracellular pathogen Shigella flexneri [27]. Regardless of the mode of delivery, HBP is a marker of bacterial activity and triggers a signaling cascade, which involves alpha-kinase 1 (ALPK1) followed by the phosphorylation-dependent oligomerization of the tumor necrosis factor (TNF-α) receptor–associated factor (TRAF)–interacting protein with the forkhead-associated domain (TIFA). These processes ultimately activate proinflammatory responses via transcriptional activation by NFκB. As the response is exclusive to Gram-negative pathogens with an ADP-heptose pathway, its presence is proposed as representing a means of differentiating between Gram-negative pathogens and the beneficial microbiota (Figure 3b).
Figure 3.
Cellular signaling with a bacterial monosaccharide.
(a) The ADP-heptose biosynthesis pathway illustrating the transketolase step, which establishes the seven-carbon skeleton. The key intermediate, HBP, is highlighted in a blue box. (b) Cellular delivery of bacterial HBP by various pathogens and signaling cascade initiated by activation of ALPK1 ultimately leading to transcriptional regulation via NF-κB stimulating an inflammatory response.
Bacterial glycan arrays
Glycan arrays are a mainstay of glycobiology research and the capabilities of this technology are being continually evolved to meet demands [28]. The arrays provide a highly efficient means of screening carbohydrate-binding proteins (GBPs) to identify their cognate carbohydrate ligands. Arrays of GBPs are also valuable and can be enlisted for the identification of glycans serving as signals in physiological interactions and responses. Glycan arrays were initially focused on mammalian glycans in recognition of the involvement of cell-surface carbohydrates in human disease [28]. However, the importance of bacterial cell-surface glycans in infection and inflammation has led to increased interest in the development and application of bacterial glycan arrays [29]. Bacterial glycan arrays may be specifically targeted to a particular genus or a species. For example, a recently-introduced array of synthetic glycans representing mycobacteria has been developed and employed to investigate interactions between the human innate and adaptive immune systems and diverse components of the glycome of Mycobacterium tuberculosis [30]. Bacterial glycan arrays, have also been developed from fractionated, but uncharacterized glycoconjugates; these are known as “shot-gun arrays” [29,31]. Although uncharacterized, these types of arrays may provide insight into carbohydrate that might be of interest in a particular type of glycoconjugate.
A new glycan array featuring linear and branched glycans (1-20 carbohydrates in size), designated as the Max Planck Society (MPS) array, has also been introduced [32]. The collection of 300 glycans includes those derived from automated and solution phase synthesis, chemoenzymatic synthesis and bacterial isolates. From this set, 140 glycans representing mammalian, protozoal and prokaryote-specific glycans has been validated in array format using plant lectins and applied to glycoimmunology and infectious disease glycobiological screens. The glycans and the metrics for the diversity of the MPS array are noted to be comparable to those from other centers. Future steps, towards the assembly of glycan arrays that represent the rich diversity of bacterial glycans will certainly contribute to a broader understanding of the implications of this diversity in human disease.
In this context, bioinformatics analyses [33], which exploit the current comprehensive glycan database of Toukach [4], and codify the most common, but unique bacterial glycans, will guide de novo and chemoenzymatic synthesis efforts to build the most relevant arrays. From these analyses it is evident that although entire prokaryotic glycomes are too extensive to screen with currently-available technologies, when arrays focus on smaller motifs such as disaccharides, the structure-space becomes much more tractable, while maintaining diversity. Other recent advances in glycan presentation also address challenges with bacterial glycan screening technologies. For example, a recent multiplex mammalian glycan bead array (MGBA) system, which is based on Luminex bead array technology, might be advantageously pivoted towards bacterial glycans [34].
Antibiotic resistance through strategic glycosylation and glycan modification
The forms and functions of bacterial glycans are even more expanded by carbohydrate modifications that occur either on saccharide building blocks prior to glycan assembly, or, by modification of assembled glycans. (See Figure 1b for a limited set of modifications.) As glycoconjugates represent the “front line” for interactions with the environment, other organisms and natural and synthetic antibiotics, these additional avenues for structural variation enable the evolution of new structures that are adapted and selected for survival under challenging conditions.
The LPS of Gram-negative bacteria contributes to structural integrity and protects the membrane from access by many small molecule antibacterial agents. The LPS features many negatively-charged functional groups including phosphoryl groups on the lipid A and heptose sugars and carboxyl groups on the Kdo sugars (Fig 1b). Alteration of this charge density, through glycan modification, affords resistance to antibiotics and antimicrobial peptides. A deceptively-simple modification, that serves to modulate glycoconjugate charge, is installation of the phosphoethanolamine (PEtN) moiety found as a site-specific modification on carbohydrates including hexoses, heptoses and Kdo in diverse glycoconjugates. Several PEtN transferases have been characterized to date. Recently two PEtN transferases representing a new family have been identified by bioinformatics analysis and characterized from Pasteurella multocida [35]. These enzymes transfer a single PEtN to Kdo in LPS and are associated with resistance to the cationic antimicrobial peptide cathelicidin-2.
Alternatively, the core peracylated β-1,6-glucosamine disaccharide diphosphate phosphate of the LPS may be modified to introduce positively-charged moieties. This is illustrated with the action of ArnT, a periplasmic glycosyl transferase that confers resistance to the cationic antibiotic polymyxin in Escherichia coli and Salmonella enterica, as well as in serious pathogens including Pseudomonas aeruginosa and Klebsiella pneumoniae [36]. Resistance results from transfer of the cationic pentose sugar, 4-amino-4-deoxy-L-α-arabinose (Ara4N, PENf, Figure 1b) from undecaprenol phosphate-α-L-Ara4N (UndP-Ara4N) to phosphoryl groups in the assembled (Kdo)2-peracylated Lipid A. This modification alters the net negative charge on the LPS and thus susceptibility to the highly-basic antibiotic. The structure of the integral membrane ArnT has recently been characterized providing mechanistic insight and details of the binding of the UndP-Ara4N and the peracylated lipid A derivative [37]. Ara4N is an unusual pentose (in pyranose form) that is biosynthesized from UDP-glucuronic acid by C4 oxidation, which facilitates C-6 decarboxylation. Then pyridoxamine-dependent transamination affords UDP-α-L-Ara4N, which is converted to UndP-Ara4N [38]. This process illustrates how the pyranose-form pentose sugars are biosynthesized from a common UDP-sugar without recourse to the metabolic pool of hexoses.
The biosynthesis of another unusual pentose, methylthio-D-xylose (MTX), has also been recently elucidated [39]. MTX originates from 5’methyl thioadenosine, a byproduct of mammalian polyamine biosynthesis. MTX is found as an α-1-4-linked modification of terminal mannoses on the lipoarabinomannan (LAM) polysaccharide in the cell envelope of Mycobacterium tuberculosis and other pathogenic mycobacteria. The uniqueness of the MTX motif has been targeted in the development of a novel immunoassay for M. tuberculosis. The assay is selective for Mycobacterium tuberculosis and shows no cross-reactivity with fast-growing mycobacteria or other bacteria [40]. The cell-surface presentation of the MTX-modified cap suggests that it plays a role in host–pathogen interactions and, although the current understanding of the function of MTX is still limited, knowledge of the biosynthetic pathway and the development of MTX-deficient M. tuberculosis strains will be critical for future studies to understand role physiological role of this rare pentose.
Conclusions
The cell-surface glycans and glycoconjugates of bacteria play essential roles in the mechanical and chemical stability of microorganisms and are intimately involved in interactions with the environment and other living organisms. In particular, the glycomes of pathogens and symbionts are of great interest as they provide opportunities for infinite structural variation to adapt to the hosts that they inhabit and thus the details of these variations are extremely important to human health.
Major progress has been made in determining the structures of complex carbohydrates through advances in biochemical, analytical and bioinformatics approaches. Despite this, current surveys suggest that the known diversity of bacterial carbohydrates, carbohydrate modifications and the glycans into which they are embedded may be just the “tip of the iceberg” and, there is the anticipation that an understanding of the sheer scope of bacterial glycomes will follow apace with advances in bacterial genome sequencing where the metrics are well-defined [41]. In addition to providing new information on glycan diversity, the surge of genomic and metagenomic data will fuel prediction of additional carbohydrate-related pathways that contribute to the biosynthesis of bacterial glycans, as genes for the biosynthesis of unique bacterial sugars are commonly clustered with those that program glycoconjugate assembly.
In the face of this progress there remain major challenges to understanding the myriad functions of carbohydrates in bacteria. As highlighted in this opinion, chemical biology approaches, including glycan arrays and metabolic oligosaccharide engineering, show great promise for defining bacterial glycomes and their interactions. In addition, the recent discovery of bacterial carbohydrates as mammalian signaling molecules and the specific roles of glycan modifications in antibiotic resistance are illustrated and suggest that there are many surprises yet to come in the Brave New World of bacterial carbohydrates.
Acknowledgements
Research on the topics reflected in this review were supported in part by the National Institutes of Health R01 GM039334. I also gratefully acknowledge the kind assistance of Dr. Philip Toukach in introducing me to the Bacterial Carbohydrate Data Base and Dr. Cristina Zamora and Prof. Karen Allen for critical editing of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The author declares no conflict of interest
References and recommended reading
Papers of particular interest published with the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Werz DB, Ranzinger R, Herget S, Adibekian A, von der Lieth CW, Seeberger PH: Exploring the structural diversity of mammalian carbohydrates (“glycospace”) by statistical databank analysis. ACS Chem Biol 2007, 2:685–691. [DOI] [PubMed] [Google Scholar]
- 2.Cummings RD: The repertoire of glycan determinants in the human glycome. Mol Biosyst 2009, 5:1087–1104. [DOI] [PubMed] [Google Scholar]
- 3.Herget S, Toukach PV, Ranzinger R, Hull WE, Knirel YA, von der Lieth CW: Statistical analysis of the Bacterial Carbohydrate Structure Data Base (BCSDB): characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct Biol 2008, 8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toukach PV, Egorova KS: Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res 2016, 44:D1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper presents the CSDB, which is accessed at: http://csdb.glycoscience.ru. The database links to the bacterial carbohydrate structure database (BCSDB), which includes the most comprehensive and powerful compilation of information on naturally-occurring bacterial carbohydrates from the scientific literature. The BCSDB is actively curated by Dr. Philip Toukach at the Zelinsky Institute.
- 5.Zunk M, Kiefel MJ: The occurrence and biological significance of the alpha-keto-sugars pseudaminic acid and legionaminic acid within pathogenic bacteria. RSC Advances 2014, 4:3413–3421. [Google Scholar]
- 6.Knirel YA, Bystrova OV, Kocharova NA, Zahringer U, Pier GB: Conserved and variable structural features in the lipopolysaccharide of Pseudomonas aeruginosa. J Endotoxin Res 2006, 12:324–336. [DOI] [PubMed] [Google Scholar]
- 7.Tytgat HL, Lebeer S: The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev 2014, 78:372–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran AP: Part I: Microbial glycolipids, glycoproteins, and glycopolymers In Microbial Glycobiology. Edited by Holst O, Brennan PJ, von Itstein M: Elsevier; 2009:1–268. [Google Scholar]
- 9.Angala SK, Palcekova Z, Belardinelli JM, Jackson M: Covalent modifications of polysaccharides in mycobacteria. Nat Chem Biol 2018, 14:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This is an excellent commentary on the diverse modifications of the mycobacterial glycome.
- 10.Dube DH, Bertozzi CR: Metabolic oligosaccharide engineering as a tool for glycobiology. Curr Opin Chem Biol 2003, 7:616–625. [DOI] [PubMed] [Google Scholar]
- 11.Keppler OT, Horstkorte R, Pawlita M, Schmidt C, Reutter W: Biochemical engineering of the N-acyl side chain of sialic acid: biological implications. Glycobiology 2001, 11:11R–18R. [DOI] [PubMed] [Google Scholar]
- 12.Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, Gagneux P, Nizet V, Varki A: Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci U S A 2009, 106:13552–13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen M, Varki A: The sialome - Far more than the sum of the parts. OMICS 2014, 14:455–464. [DOI] [PubMed] [Google Scholar]
- 14.Laughlin ST, Bertozzi CR: Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat Protoc 2007, 2:2930–2944. [DOI] [PubMed] [Google Scholar]
- 15.Schoenhofen IC, McNally DJ, Brisson JR, Logan SM: Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 2006, 16:8C–14C. [DOI] [PubMed] [Google Scholar]
- 16.Schoenhofen IC, Vinogradov E, Whitfield DM, Brisson JR, Logan SM: The CMP-legionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 2009, 19:715–725. [DOI] [PubMed] [Google Scholar]
- 17.Holden HM, Cook PD, Thoden JB: Biosynthetic enzymes of unusual microbial sugars. Curr Opin Struct Biol 2010, 20:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andolina G, Wei R, Liu H, Zhang Q, Yang X, Cao H, Chen S, Yan A, Li XD, Li X: Metabolic labeling of pseudaminic acid-containing glycans on bacterial surfaces. ACS Chem Biol 2018, 13:3030–3037. [DOI] [PubMed] [Google Scholar]; ** Studies on the synthesis and applications of azide analogs of an intermediate in pseudaminic acid biosynthesis are presented. By using an advanced intermediate in the biosynthesis authors elegantly demonstrate the value of MOE for selective live-cell imaging of the LPS of selected strains of the human pathogen P. aeruginosa.
- 19.Mas Pons J, Dumont A, Sautejeau G, Fugier E, Baron A, Dukan S, Vauzeilles B: Identification of living Legionella pneumophila using species-specific metabolic lipopolysaccharide labeling. Angew Chem Int Ed Engl 2014, 53:1275–1278. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson I, Grove K, Dovala D, Uehara T, Lapointe G, Six DA: Molecular characterization and verification of azido-3,8-dideoxy-D-manno-oct-2-ulosonic acid incorporation into bacterial lipopolysaccharide. J Biol Chem 2017, 292:19840–19848. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Selective metabolic labeling of core Kdo saccharides in LPS is reported and although studies clearly confirm incorporation, the authors provide important information on the efficiency of the Kdo-N3 precursor relative to Kdo, which must be taken into consideration in experimental design.
- 21.Wang W, Zhu Y, Chen X: Selective imaging of Gram-negative and Gram-positive microbiotas in the mouse gut. Biochemistry 2017, 56:3889–3893. [DOI] [PubMed] [Google Scholar]; ** This study illustrates how the dual application of MOE and a fluorescent Vancomycin deriative enables simultaneous selective imaging of Gram-negative and Gram-positive microbiota in the mouse gut - highlighting the power of chemical tools in very complex applications.
- 22.Nilsson I, Prathapam R, Grove K, Lapointe G, Six DA: The sialic acid transporter NanT is necessary and sufficient for uptake of 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) and its azido analog in Escherichia coli. Mol Microbiol 2018, 110:204–218. [DOI] [PubMed] [Google Scholar]
- 23.Mahla RS, Reddy MC, Prasad DV, Kumar H: Sweeten PAMPs: Role of sugar complexed PAMPs in innate immunity and vaccine biology. Front Immunol 2013, 4: Article 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudet RG, Sintsova A, Buckwalter CM, Leung N, Cochrane A, Li J, Cox AD, Moffat J, Gray-Owen SD: Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science 2015, 348:1251–1255. [DOI] [PubMed] [Google Scholar]
- 25.Kneidinger B, Marolda C, Graninger M, Zamyatina A, McArthur F, Kosma P, Valvano MA, Messner P: Biosynthesis pathway of ADP-L-glycero-beta-D-manno-heptose in Escherichia coli. J Bacteriol 2002, 184:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gall A, Gaudet RG, Gray-Owen SD, Salama NR: TIFA Signaling in gastric epithelial cells initiates the cag Type 4 secretion system-dependent innate immune response to Helicobacter pylori Infection. MBio 2017, 8:e0116816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaudet RG, Guo CX, Molinaro R, Kottwitz H, Rohde JR, Dangeard AS, Arrieumerlou C, Girardin SE, Gray-Owen SD: Innate recognition of intracellular bacterial growth Is driven by the TIFA-dependent cytosolic surveillance pathway. Cell Rep 2017, 19:1418–1430. [DOI] [PubMed] [Google Scholar]; ** A new role for bacterial carbohydrates is presented in this and related papers on the action of heptose bisphosphate (HBP). This most recent paper shows how a simple bacterial metabolite, HBP, which is released during cytosolic growth of intracellular pathogens such as S. flexnieri, ultimately triggers a signaling pathway that amplifies the in flammatory response to virulent pathogens.
- 28.Rillahan CD, Paulson JC: Glycan microarrays for decoding the glycome. Annu Rev Biochem 2011, 80:797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stowell SR, Arthur CM, McBride R, Berger O, Razi N, Heimburg-Molinaro J, Rodrigues LC, Gourdine JP, Noll AJ, von Gunten S, et al. : Microbial glycan microarrays define key features of host-microbial interactions. Nat Chem Biol 2014, 10:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng RB, Jegouzo SAF, Joe M, Bai Y, Tran HA, Shen K, Saupe J, Xia L, Ahmed MF, Liu YH, et al. : Insights into interactions of mycobacteria with the host innate immune system from a novel array of synthetic mycobacterial glycans. ACS Chem Biol 2017, 12:2990–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes a glycan array representing the major carbohydrate structures present in the cell wall of the Mycobacterium tuberculosis and other mycobacteria. The power of the array is demonstrated by investigating interactions between mycobacterial glycans and receptors that mediate uptake and survival of Mtb in macrophages, dendritic cells, and sinusoidal endothelial cells.
- 31.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD, Smith DF: Shotgun glycomics: A microarray strategy for functional glycomics. Nat Methods 2011, 8:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geissner A, Reinhardt A, Rademacher C, Johannssen T, Monteiro J, Lepenies B, Thepaut M, Fieschi F, Mrazkova J, Wimmerova M, et al. : Microbe-focused glycan array screening platform. Proc Natl Acad Sci U S A 2019, 116:1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper introduces the development and applications of a new glycan array, which includes many prokaryote-specific glycans from diverse organisms.
- 33.Adibekian A, Stallforth P, Hecht M-L, Werz DB, Gagneux P, SP H: Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem Sci 2011, 2:337–344. [Google Scholar]
- 34.Purohit S, Li T, Guan W, Song X, Song J, Tian Y, Li L, Sharma A, Dun B, Mysona D, et al. : Multiplex glycan bead array for high throughput and high content analyses of glycan binding proteins. Nat Commun 2018, 9: Article 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper M, Wright A, St Michael F, Li J, Deveson Lucas D, Ford M, Adler B, Cox AD, Boyce JD: Characterization of two novel lipopolysaccharide phosphoethanolamine transferases in Pasteurella multocida and their role in resistance to cathelicidin-2. Infect Immun 2017, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olaitan AO, Morand S, Rolain JM: Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 2014, 5:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrou VI, Herrera CM, Schultz KM, Clarke OB, Vendome J, Tomasek D, Banerjee S, Rajashankar KR, Belcher Dufrisne M, Kloss B, et al. : Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science 2016, 351:608–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raetz CR, Reynolds CM, Trent MS, Bishop RE: Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem 2007, 76:295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angala SK, McNeil MR, Shi L, Joe M, Pham H, Zuberogoitia S, Nigou J, Boot CM, Lowary TL, Gilleron M, et al. : Biosynthesis of the methylthioxylose capping motif of lipoarabinomannan in Mycobacterium tuberculosis. ACS Chem Biol 2017, 12:682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigal GB, Pinter A, Lowary TL, Kawasaki M, Li A, Mathew A, Tsionsky M, Zheng RB, Plisova T, Shen K, et al. : A Novel Sensitive Immunoassay Targeting the 5-Methylthio-D-Xylofuranose-Lipoarabinomannan Epitope Meets the WHO’s Performance Target for Tuberculosis Diagnosis. J Clin Microbiol 2018, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, et al. : Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 2015, 15:141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]