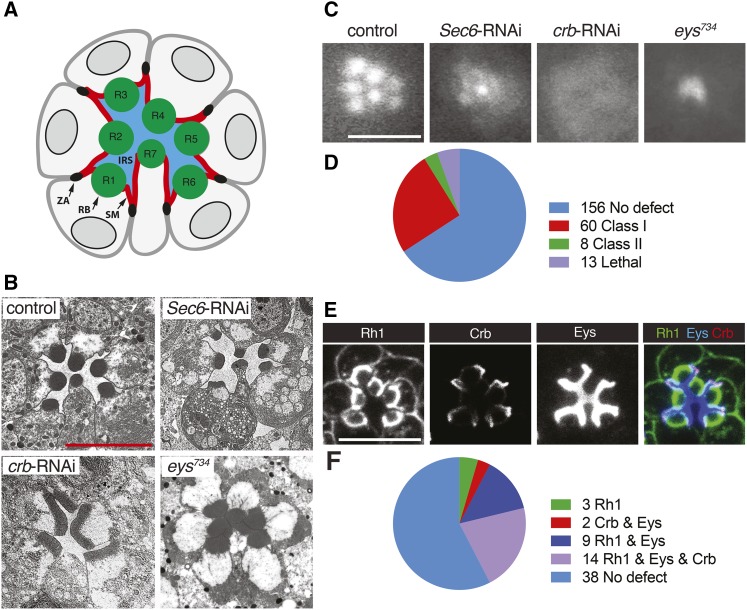
Figure 2.
RNAi-based screen of known and predicted trafficking factors in Drosophila PRCs. GMR-GAL4 was used to drive the expression of UAS-dicer-2 and UAS-RNAi constructs. UAS-dicer-2/+; pGMR-Gal4/+ was used as control. Scale bars, 5 μm. (A) Schematic of cross-section of PRCs in a Drosophila ommatidium. Apical membranes of PRCs are subdivided into the rhabdomere (RB) and stalk membrane (SM). The stalk membrane connects the rhabdomere to the zonula adherens (ZA). PRCs R1-R8 (R8 is found below R7) surround the interrhabdomeral space (IRS). (B) TEM images of a wild-type, Sec6-RNAi, crb-RNAi, and eys734 mutant ommatidium. (C) TLI images of control, Sec6-RNAi, crb-RNAi, and eys734 mutant ommatidium. (D) Summary of the TLI screen. (E) Distribution of Rh1, Crb, and Eys in ommatidia of control flies. (F) Summary of data obtained from Rh1, Crb, and Eys retinal immunostaining of 66 Class I and Class II candidates identified with TLI (see also Figures S1-S4). Knockdown of 3 genes changed the distribution of Rh1, knockdown of 2 genes changed the distribution of both Crb and Eys, knockdown of 9 genes changed the distribution of both Rh1 and Eys, and knockdown of 14 genes changed the distribution of Rh1, Crb, and Eys.