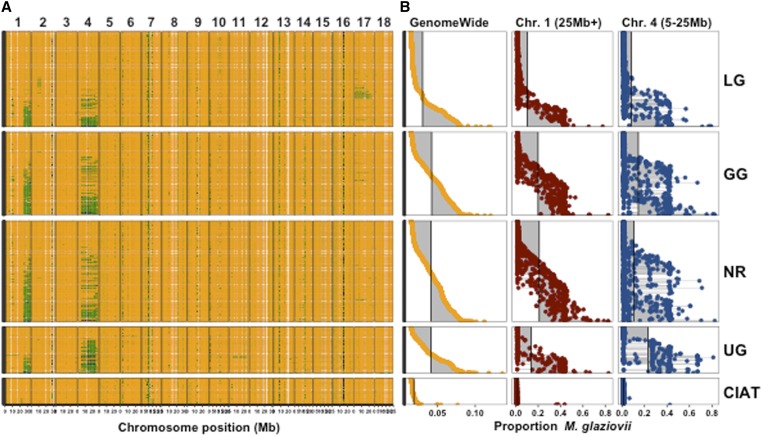
Figure 2.
Comparison of introgression among populations. The mean M. glaziovii allele dosage at IDMs in 250 kb windows across the genome is depicted on the left (A). Physical position on each chromosome is depicted in megabases (Mb) along the x-axis. Colors range from orange (0 M.g. alleles), to green (1 M.g. allele), to dark blue (2 M.g. alleles). The per individual proportion of M. glaziovii alleles at IDM is summarized on the right (B). Proportions were calculated as the sum (per clone) of the dosages at IDMs divided by two times the number of IDMs. The proportions in (B) were computed either with all IDMs (GenomeWide, left column), at IDMs on chr. 1 >25 Mb (middle column), at IDMs on chr. 4 from 5 to 25 Mb (right column). The populations can be compared in (B) by looking down the columns and using the vertical lines, which represent the mean values for that group and region, as a visual aid. For both (A and B) each row (y-axis) is an individual cassava clone and the vertical panels represent five populations: IITA local germplasm (LG), IITA Genetic Gain (GG), NRCRI (NR), NaCRRI (UG) and the L. American collection (CIAT). Rows (clones) are aligned across (A and B) and sorted within population based on the genome-wide proportion M. glaziovii [left column of (B)].