Abstract
Stem cell systems regulate tissue development and maintenance. The germline stem cell system is essential for animal reproduction, controlling both the timing and number of progeny through its influence on gamete production. In this review, we first draw general comparisons to stem cell systems in other organisms, and then present our current understanding of the germline stem cell system in Caenorhabditis elegans. In contrast to stereotypic somatic development and cell number stasis of adult somatic cells in C. elegans, the germline stem cell system has a variable division pattern, and the system differs between larval development, early adult peak reproduction and age-related decline. We discuss the cell and developmental biology of the stem cell system and the Notch regulated genetic network that controls the key decision between the stem cell fate and meiotic development, as it occurs under optimal laboratory conditions in adult and larval stages. We then discuss alterations of the stem cell system in response to environmental perturbations and aging. A recurring distinction is between processes that control stem cell fate and those that control cell cycle regulation. C. elegans is a powerful model for understanding germline stem cells and stem cell biology.
Keywords: stem cell, niche, physiology, network, Notch, WormBook, Caenorhabditis elegans
General Features of the Caenorhabditis elegans Germline Stem Cell System
The Caenorhabditis elegans germline stem cell system displays several features in common with stem cell systems from a variety of other animals and other organs. We list 10 such features here that we take up in more detail in the context of the review.
Renewal and differentiation: C. elegans germline stem cells exhibit the two defining features of stem cells, self-renewal and generation of cellular progeny that differentiate. C. elegans germline stem cells generate a large number of gametes over an extended portion of adult life (Hirsh et al. 1976).
Niche: Like the majority of stem cell systems characterized to date, C. elegans germline stem cells require interaction with a local stem cell “niche.” In fact, a functional stem cell niche— a local microenvironment of a different cell type than the stem cells, outside of which stem cells differentiate—was first demonstrated in pioneering laser microsurgery experiments by Kimble and White in 1981 (Kimble and White 1981), just 3 years after Schofield put forward the niche hypothesis (Schofield 1978) (Figure 1).
Notch: C. elegans germline stem cells require Notch signaling. The niche provides ligands that activate a Notch family receptor, GLP-1, on the surface of the nearby germ cells (Hansen and Schedl 2013; Kershner et al. 2013). The Notch pathway also plays a prominent role in mammalian stem cell regulation, including those of the neural, muscle, intestinal, mammary, bone, and hematopoietic lineages where, depending on the context and developmental stage, it promotes stem cell maintenance or differentiation (Liu et al. 2010; Koch et al. 2013) (Figure 2).
Population renewal: self-renewal of C. elegans stem cell pool appears to occur at the population level, as in many mammalian stem cell systems (Morrison and Kimble 2006). This contrasts with the well-characterized lineal asymmetric cell renewal mechanism characteristic of Drosophila germline stem cells (Fuller and Spradling 2007) (Figure 3).
No defined lineage: at the cellular level, germ cell divisions do not follow a defined lineage. This feature contrasts with somatic cells in C. elegans that follow an invariant developmental lineage from animal to animal and allowed the complete somatic cell lineage to be determined (Sulston and Horvitz 1977; Sulston et al. 1983). Also, as in many mammalian stem cell systems, C. elegans germ cells divide asynchronously.
Multipotential: the stem cells are undifferentiated but multipotential. Because of the nature of C. elegans hermaphroditism, germline stem cells generate first male and then female gametes in the same individual.
Quiescence and activation: the stem cells can be either mitotically cycling or quiescent, depending on conditions. When dividing, C. elegans germline stem cells display special cell cycle features including an extremely short G1. Drosophila germline stem cells also have a short G1 and long G2 (Hsu et al. 2008). When quiescent, C. elegans germline stem cells arrest in the G2, similar to embryonic germ cells and a subset of neural stem cells in Drosophila (Su et al. 1998; Otsuki and Brand 2018).
Regeneration: C. elegans germline stem cells can regenerate the entire germ line, including all developmental intermediates, from a pool of stem cells. In a particularly dramatic example of complete starvation in adults, the developed germ line is resorbed, save a small pool of stem cells that can regenerate the entire germ line and restore fertility under appropriate conditions (Angelo and Van Gilst 2009). Like several other stem cell systems, germ cells also exhibit more subtle plasticity, such as modulation of the progenitor pool by conserved signaling mechanisms that respond to organismal physiological conditions (Ables et al. 2012; Hubbard et al. 2013; Laws and Drummond-Barbosa 2017).
Adult activity: C. elegans germline stem cells divide in the adult animal. No other cells divide in the adult.
Depletion with age: like other stem cell systems (Drummond-Barbosa 2008; Oh et al. 2014; Schultz and Sinclair 2016), C. elegans germline stem cell numbers decline with age, as does the pace of cell cycle progression.
Figure 1.
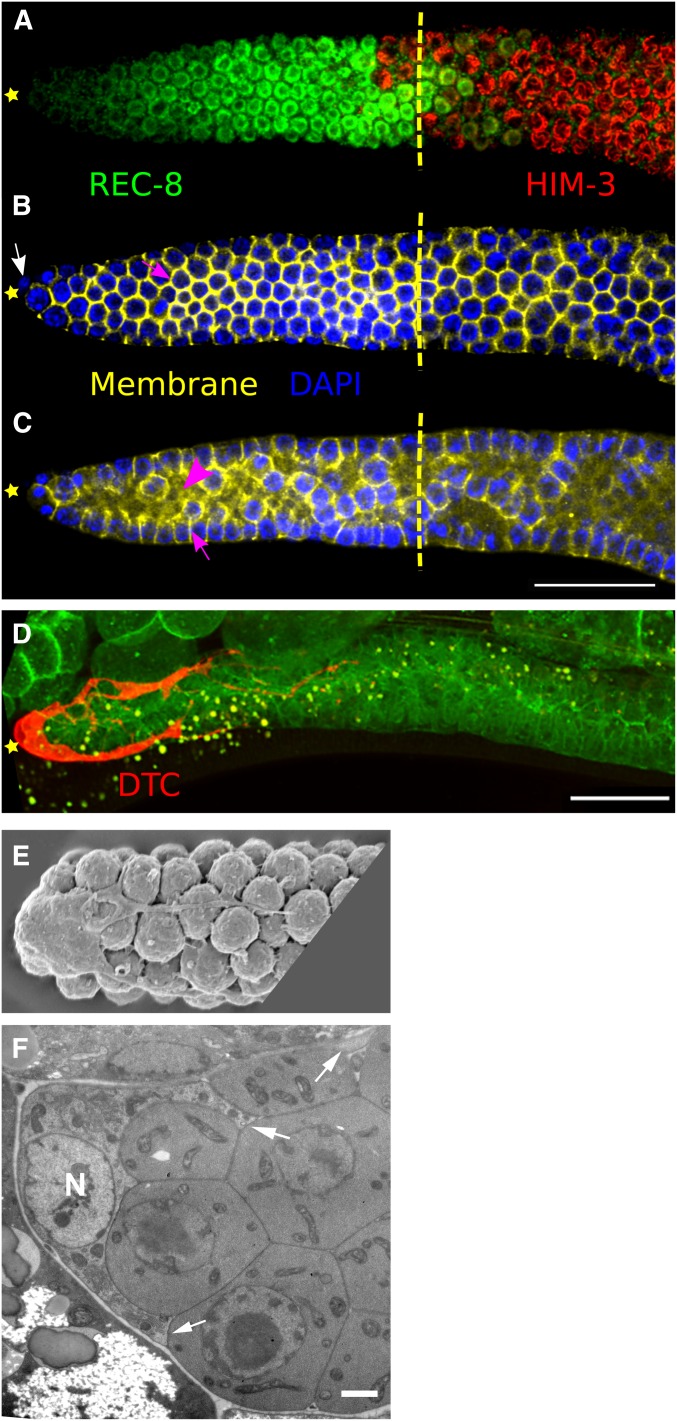
Cytology of the germline stem cell system. (A–C) Fluorescence micrographs of a dissected young adult hermaphrodite gonad. (A) Progenitor zone (PZ) cells are marked in green (nucleoplasmic REC-8) and cells in leptotene-zygotene are marked in red (meiotic chromosome axis protein HIM-3). (B) Surface view and (C) internal view of gonad with cell plasma membranes in yellow [GFP::PH(PLCdelta)] and nuclei in blue (DAPI). Pink arrows and arrowheads indicate plasma membranes and core (or rachis), respectively. For all panels, distal is left and proximal is right; white arrow marks distal tip cell (DTC) nucleus, yellow star marks the distal tip of the gonad, and yellow vertical dashed line marks the boundary of the PZ and leptotene. In this gonad, the boundary is at cell diameter 21 from the distal tip where more half of the cells in a row have switched from strong nucleoplasmic REC-8 staining to HIM-3 staining. (D) Fluorescence micrograph of DTC cytoplasm in a live young adult hermaphrodite (red, lag-2p::mCherry) and membranes (green, SYN-4::GFP). Bar, 20 µM (A–D). (E) Scanning electron micrograph of the surface of a dissected young adult hermaphrodite gonad (Hall et al. 1999). The dissected gonad preparation was digested with proteases prior to fixation, highlighting the DTC body, surface germ cells, and some DTC processes; removed are the surrounding basement membrane, intercalating DTC membranes and many processes. (F) Transmission electron micrograph of an interior section of a young adult hermaphrodite; DTC nucleus (N); arrows indicate intercalating DTC membranes and cytoplasm. Bar, 1 µM. Despite a small opening to the central core, each germ cell nucleus as seen in A–C and F, is surrounded by its own plasma membrane and cytoplasm as seen in E and F. A–C are from Ariz Mohammad, D is from Olga Pekar, and E and F are from David Hall (Hall et al. 1999), with permission.
Figure 2.
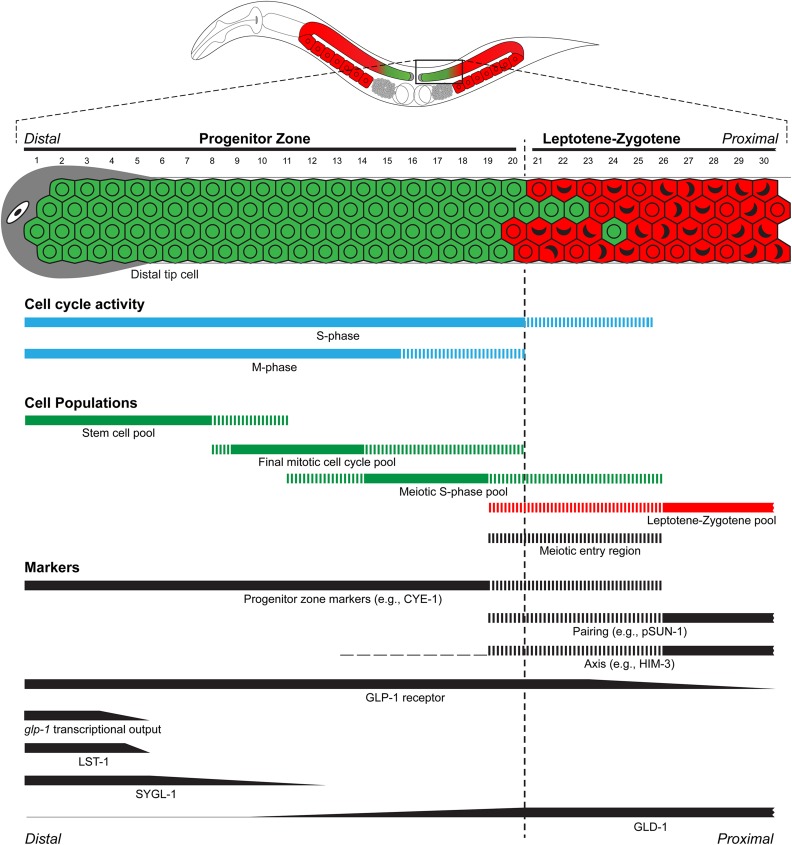
Organization and markers in the germline stem cell system. Schematic diagram of the distal germ line and the approximate extent of cell pools and marker accumulation as observed in the “day 1” adult hermaphrodite (∼24 hr past the mid-L4) under standard laboratory conditions (see text for details and references). Distal tip cell (gray), PZ cells (green), and leptotene-zygotene cells (red). Cell diameter numbers are indicated, with one at the distal tip through 30 in zygotene. The vertical black dashed line indicates the boundary of the PZ and leptotene (corresponding to yellow vertical line in Figure 1). The extent of M phase and S phase cell cycle activity is shown in blue horizontal bar, based on EdU incorporation and phospho-H3 staining, respectively. Cell populations in the progenitor zone (PZ; green bars): the stem cell pool, final mitotic cell cycle pool, and meiotic S phase pool as inferred from cell population analysis. The leptotene-zygotene pool (red bar) is based on staining of meiotic chromosome pairing or axis. Marker gene products (black bars): PZ markers include CYE-1, REC-8, and WAPL-1. Leptotene/overt meiotic entry markers include gene products that participate in meiotic chromosome pairing (e.g., phospho-SUN-1) or are part of the meiotic chromosome axes (e.g., HIM-3). For categorical markers and activities that show nuclear staining (PZ markers, etc.), the solid bar indicates region where all cells stain, while vertical hatching indicates region where only a subset of cells have nuclear staining. For meiotic prophase marker gene products whose accumulation is repressed by FBF (e.g., HIM-3, see text), accumulation is observed in the cytoplasm of the proximal PZ, as indicated by horizontal dashes. In the meiotic entry region (black vertical hatching) cells stain with either PZ or leptotene markers. Accumulation of regulator proteins GLP-1, LST-1, SYGL-1, and GLD-1 is based on antibody staining, while glp-1 transcriptional output is based on single-molecule fluorescent in situ hybridization using intron probes for lst-1 and sygl-1.
Figure 3.
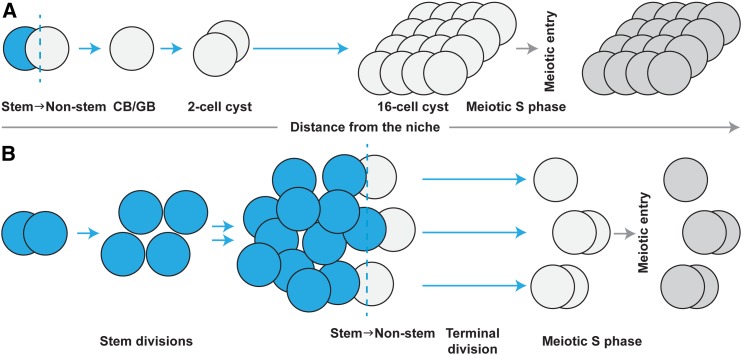
Comparison of single stem cell asymmetry and population asymmetry in germline stem cell systems of Drosophila and C. elegans. Schematic representation of the Drosophila and C. elegans adult germline stem cell systems, focusing on germ cell behavior. (A) Model for the Drosophila adult germline stem cell behavior. An asymmetric germline stem cell division gives rise to a daughter that is displaced from niche and becomes a nonstem progenitor (termed cystoblast “CB” in female and gonialblast “GB” in male) that synchronously divides four times to give rise to 16 cells, one of which becomes the oocyte in the female (the other 15 become nurse cells) and all of which become spermatocytes in the male. The latter divisions are considered “transit amplifying” divisions since many cells are generated for each stem/nonstem division. This renewal strategy can maintain a tissue with a small number of stem cells, but requires multiple cell divisions (and time) to regenerate the full complement of differentiating progeny. (B) Model for C. elegans adult hermaphrodite germline stem cell behavior. A symmetric stem cell division (within the pool of stem cells) gives rise to two stem cells, either or both of which may remain distal or may be displaced from the distal-most region, but each of which undergo, on average, an additional 1–2 symmetric germline stem cell divisions. Stem daughters are not maintained in a cyst, do not divide synchronously, and may be separated from each other; stem and nonstem pools of cells overlap to some extent (see Figure 2). After falling below a critical level of response to niche signaling (GLP-1 Notch), cells (three shown) become nonstem cells (indicated in gray), complete their ongoing mitotic cell cycle before entering meiotic S phase, and reach overt meiotic prophase (leptotene). No “transit amplifying” divisions of the nonstem daughter occur in this model. This renewal strategy requires a relatively large stem cell population but a relatively small number of cell divisions (and therefore relatively little time) to regenerate the full complement of differentiating progeny. Note that somatic gonad cells are not depicted.
The combination of these conserved features in a fast-growing, highly fecund, optically clear model organism that is eminently accessible to genetic and molecular manipulation (Corsi et al. 2015) enhances its power as a model for stem cell biology.
The germline stem cell system in C. elegans, as described in detail below, includes the distal tip cell (DTC) niche and the progenitor zone (PZ), where the PZ includes stem cells, progenitor cells (corresponding to cells completing their ongoing mitotic cell cycle), and cells in meiotic S phase. The majority of the past work on the stem cell system focuses on the PZ as a whole. Recent work identified gene products that can mark stem cells, and these will help us to further understand and dissect the stem cell system going forward.
A major approach to understanding the C. elegans germline stem cell system is genetics. Genetic approaches that have yielded key regulators of the stem cell system include the identification and characterization of (i) mutations and RNA interference (RNAi) knockdowns that cause abnormalities in the stem cell system, (ii) genetic modifiers (e.g., suppressors and enhancers) of these phenotypes, and (iii) their functional relationships. Genetically identified regulators include genes that promote the stem cell fate (notably Notch pathway components), cell cycle progression, and differentiation, and that mediate the effects of physiology on the system.
Historically, C. elegans germline transgene expression lagged nearly two decades behind somatic multicopy transgene expression technology. Ironically, this barrier in germline transgene expression indirectly led to the Nobel Prize winning identification of RNAi (Guo and Kemphues 1995; Fire et al. 1998). Thanks to technical advances, which enable routine single-copy insertions and precise genome editing such as MosSCI and CRISPR/Cas9 genome editing, germline gene manipulations are now routine (Nance and Frøkjær-Jensen 2019). The ability to tag germline proteins in their native genomic context is a particular boon.
Two general processes control the stem cell system: cell fate choice (that is, the decision between the undifferentiated stem/progenitor fate vs. differentiation into the meiotic developmental pathway) and cell cycle regulation. Essentially, only cells in the undifferentiated state are competent to proliferate, and their rate of proliferation is controlled independently. Once cells have entered the meiotic pathway, they are normally no longer competent to undergo mitotic proliferation. A central issue in understanding the C. elegans germline stem cell system under diverse conditions is determining whether an alteration in cell number, dynamics, or functional output arises from a change in cell fate choice, a change in mitotic cell cycle activity, or both. These are key distinctions that we will return to throughout the review.
Below we summarize several salient features of C. elegans biology. Since the vast majority of work on the C. elegans germline stem cell system has been done in the hermaphrodite, we focus our attention on that work. For a recent, comprehensive study of the germline stem cell system in males, see Crittenden et al. (2019). Here, we first describe the cell biology and molecular regulation of cell division and stem cell fate in the adult, including a brief comparison of the C. elegans and Drosophila germline stem cell systems. We then turn to the development of the stem cell system during larval stages, its response to environmental challenges, and its decline with age. Finally, we consider the major outstanding questions in the field.
Overview of Salient Features of C. elegans Development, Reproduction, and Ecology
As in many other animals, the C. elegans germline lineage is set aside from somatic cells during embryogenesis. In C. elegans, it occurs over the course of the first four cell divisions. The sole germline blastomere (P4) then divides to produce the primordial germ cells (PGCs), called Z2 and Z3. During embryogenesis, these two cells are then joined by the migrating somatic gonad precursors (SGPs) Z1 and Z4, and these four “Z” cells form the gonad primordium, a structure easily seen in late embryonic stages and in the hatchling, surrounded by a basement membrane in which all subsequent germline and somatic gonad development occur (Kimble and Hirsh 1979; Sulston et al. 1983).
In standard laboratory conditions, the hatchling continues development from the first larval stage (L1) through three additional larval stages (L2–L4) followed by the reproductively mature adult stage. A molt punctuates each stage transition, and the entire egg-to-egg cycle takes 3–4 days at 20°. Germ cells remain mitotically quiescent through the first half of the L1 but, provided the animal feeds, germ cells begin to proliferate about midway through the L1 stage. At the L2/L3 transition in the hermaphrodite, a reorganization of the somatic gonadal cells (12 descendants of Z1 and Z4) bisects the growing germ cell population and segregates cells into anterior and posterior “arms” of the developing hermaphrodite gonad (Kimble and Hirsh 1979). At the mid-L3, “initial meiosis” occurs. This event is defined as the earliest time in development when germ cells enter the meiotic pathway. Initial meiosis establishes the pattern of distal germline stem/progenitor cells relative to the more proximal germ cells that are in increasingly mature stages of meiosis and gametogenesis along the distal to proximal axis (Figure 2, top). The stem/progenitor pool continues to expand in the L3–L4 stages, establishing an adult pool that is maintained during early adulthood and subsequently declines.
C. elegans individuals are either hermaphrodite or male. Hermaphrodites are essentially females (sharing the same basic gonadal plan of females of related species that are obligate females and males) that produce sperm in the last larval stage (L4). These sperm, which reside in the spermatheca after the first ovulation in the adult, then fertilize the continuous supply of oocytes that are produced in the adult. The number of self-sperm limits the number of progeny hermaphrodites produce without mating such that wild-type animals under laboratory conditions typically produce ∼300 progeny, from ∼35 to 40 male germ cells in each gonad arm, which each give rise to four haploid sperm after completion of meiosis. After the spermatogenesis/oogenesis switch (Zanetti and Puoti 2013), germ cells that enter the meiotic pathway produce oocytes or serve as nurse cells that are culled by cell death. Oocytes line up assembly-line fashion in the proximal gonad and, in the presence of sperm, undergo meiotic maturation one by one, after which they are each ovulated into the spermatheca, fertilized, and proceed to the uterus (Huelgas-Morales and Greenstein 2018). After early embryogenesis in the uterus, embryos are expelled from the animal, completing embryonic development and hatching outside of the mother. Thus, the flux of germ cells in and out of the system is governed by the rates of distal stem/progenitor cell production and meiotic entry on the distal side, the rates of oocyte meiotic maturation and ovulation on the proximal side, and the rate of meiotic progression and gametogenesis in between. Since oocyte maturation and ovulation occur in response to hormonal signals from sperm, in the absence of sperm—either due to genetic disruption of the germline sex determination pathway or the depletion of stored sperm over time—meiotic maturation and ovulation slows markedly (a 10–100 times drop in maturation/ovulation per hr; McCarter et al. 1999). If hermaphrodites mate with a male, even after self-sperm depletion, the abundance of sperm deposited into the hermaphrodite can support continued meiotic maturation and the production of upward of 1000 progeny.
Representations of the process of germline development over time can be found in a video cartoon on WormAtlas (https://www.wormatlas.org/hermaphrodite/germ%20line/Germframeset.html) and a run of a computational model of germline development [see Supplement to Atwell et al. (2015)].
While C. elegans has been raised and studied in the laboratory for over 50 years, its ecology has only recently attracted scrutiny (Schulenburg and Félix 2017). Since the germ line is responsible for survival of the species, the animal’s ecology is of primary relevance to the germ line. In the wild, C. elegans thrives in environments with patchy “boom and bust” resources, such as bacteria growing on rotting apples (Frézal and Félix 2015). Prior to the developmental commitment to reproduction, animals faced with dwindling resources can, thanks to developmental plasticity mechanisms, strategically delay reproduction until a more hospitable environment is encountered. The best-studied delay point is dauer, an alternate L2–L3 larval stage that is adapted to survive harsh conditions and during which germline progenitor cell production is reversibly arrested. Larvae that are too old to enter dauer can nevertheless limit germline progenitor accumulation in response to declining conditions and thereby limit reproductive output (Hubbard et al. 2013). One view is that the tremendous developmental plasticity displayed by this species in the face of adverse conditions is primarily for the success, however limited, of the germ line to produce progeny, and to produce them in numbers appropriate for the environmental condition.
Adult Germline Stem Cell System
The germline stem cell system has three phases in wild-type hermaphrodites under optimal laboratory growth conditions: larval expansion, young adult homeostasis, and age-related decline. The young adult (∼12–60 hr post L4/adult molt, 20°) is the period of maximum progeny production (peaking at ∼150/day) and the germline stem cell system is considered to be at steady state where stem/progenitor cell number, mitotic cell cycling activity, and the production of differentiating meiotic germ cells is largely constant. During this time, a balance likely exists between stem/progenitor cell production and meiotic entry. This period of young adult homeostasis has been studied in greatest detail and is presented in this section. In subsequent sections, we cover larval expansion and age-related decline.
Adult germline stem cell system: organization and cell biology
We first describe the overall organization of the adult hermaphrodite germ line and features of cells that constitute the germline stem cell system. The adult hermaphrodite has one anterior and one posterior, U-shaped gonad. Each gonad is a tube-shaped organ containing ∼1000 germ cells organized as a distal-to-proximal assembly line displaying germline development in spatiotemporal order. At the distal end is a population of mitotically cycling cells that, upon reaching a more proximal position, enter and progress through meiotic prophase and oogenesis, with the most proximal oocyte undergoing meiotic maturation, ovulation, and fertilization. The germline stem cell system and early meiotic prophase are shown in dissected gonad preparations, a live specimen, electron micrographs, and schematically in Figure 1 and Figure 2. At the distal end of the germ line is the PZ region (also called the mitotic region or proliferative zone), consisting of 200–250 germ cells and extending ∼20 cell diameters from the distal tip. The PZ contains stem and progenitor cells that are mitotically cycling based on observing M phase and S phase throughout the region (Hansen et al. 2004a; Crittenden et al. 2006; Maciejowski et al. 2006; Jaramillo-Lambert et al. 2007; Fox et al. 2011; Seidel and Kimble 2015). Following the PZ, germ cells enter leptotene of meiotic prophase and progress to zygotene more proximally. The region of the gonad containing germ cells in these early stages of meiotic prophase is also called the “transition zone.” Cells in this region are differentiating and are no longer part of the proliferating population of germ cells. These are not to be confused with “transit” cells in other stem cell systems.
Molecular markers and chromosome morphology distinguish cells in the PZ from cells in early stages (leptotene-zygotene) of meiotic prophase (Figure 1 and Figure 2). PZ cell-specific markers include proteins associated with mitotic cell cycle activity, such as CYE-1 (cyclin E) and KNL-2 (kinetochore protein), as well as WAPL-1 (cohesin chaperone) and nucleoplasmic REC-8. These latter proteins, although associated with meiosis, mark progenitor cells under certain mild fixation conditions (Hansen et al. 2004a; Fox et al. 2011; Mohammad et al. 2018). Proteins that mark leptotene-zygotene are associated with the execution of the earliest processes of meiotic prophase (Hillers et al. 2017), including homologous chromosome pairing (phosphorylated forms of SUN-1 and pairing center proteins HIM-8 and ZIM-1, -2, and -3), formation of meiotic chromosome axes (HIM-3), and sister chromatid cohesion (COH-3 and -4); these markers are essentially absent from nuclei in the PZ (Zetka et al. 1999; Penkner et al. 2009; Severson and Meyer 2014; Kim et al. 2015). Chromosome morphology, as revealed by DAPI staining (or by histone-reporter fusion proteins), changes dramatically in meiotic prophase. As a consequence of homolog pairing during leptotene, the chromatin and nucleolus are spatially reorganized resulting in a crescent-shaped DAPI morphology, which is a convenient indicator of meiotic entry (MacQueen and Villeneuve 2001). However, the crescent-shaped DAPI morphology is not observed in all wild-type leptotene cells, nor in mutants that exhibit defective pairing, show overproliferation, or have abnormal nuclear morphology (Fox and Schedl 2015; Mohammad et al. 2018). Therefore, marker staining is the preferred method for identifying PZ or meiotic prophase cells at single-cell resolution.
In young adults, germ cells leave the PZ and enter leptotene over an ∼8 cell diameter zone called the meiotic entry region (Figure 2; Hansen et al. 2004a). Since this boundary is not sharp in the adult, several operational definitions for the position of meiotic entry have been adopted. For DAPI-stained preparations in which germline nuclear morphology is regular and distal-proximal pattern is intact, the trained eye can distinguish between crescent-shaped meiotic prophase nuclei and metaphase or anaphase nuclei, and the border of meiotic entry is operationally defined as the row of cells in which two or more crescent-shaped nuclei appear (Crittenden et al. 2006). The appearance of these nuclei is followed, proximally, by rows in which additional crescent-shaped nuclei appear. For the single-cell markers described above, it is the point where more than half of the cells in a row have switched from strong staining with a PZ marker to a leptotene marker.
From the distal tip through late pachytene, germ cells are largely found on the surface of the gonad tube. Each germ cell contains a ring channel or intracellular bridge (Matova and Cooley 2001) on the interior surface that connects to an acellular region (called the rachis or core) (Hirsh et al. 1976; Hall et al. 1999; Maddox et al. 2005; Amini et al. 2014). When germ cells divide, the spindle is typically oriented in parallel to the surface of the gonad tube and the ring channel is bifurcated during cytokinesis, distributing one to each daughter on the plasma membrane facing the rachis (Seidel et al. 2018); this process appears similar to that observed in germ cell divisions in annelids (Swiatek et al. 2009) and may also share features with Drosophila pole cell divisions (Cinalli and Lehmann 2013), although the exact mechanism by which germ cells undergo cytokinesis in C. elegans while remaining attached to the rachis is unknown. Because of the ring-channel openings to the core, the PZ is technically a syncytium. However, each germ cell nucleus and cytoplasm are surrounded by their own cell plasma membranes and function as individual germ cells (Figure 1). Moreover, PZ cells cycle asynchronously, suggesting they do not share cytoplasmic cell cycle regulatory components, and neighboring cells can display differential marker expression (e.g., Gerhold et al. 2015; Lee et al. 2016; Shin et al. 2017; Mohammad et al. 2018), indicating that RNA and protein regulators are generally restricted to individual cells. Additionally, the flow of contents within the acellular region is predominantly, if not exclusively, distal-to-proximal, toward developing oocytes (Wolke et al. 2007). Thus the PZ is unlike the syncytial early Drosophila blastoderm or cells within Drosophila germline cysts that divide synchronously and share cytoplasmic contents (see Spradling et al. 2011). The PZ also contains local interior germ cells that partially span the rachis, resulting in a zig-zag organization in the rachis (Cinquin et al. 2015; Gopal et al. 2017). The position of the interior germ cells differs from gonad to gonad and within a single gonad it changes over time. The current model is that interior germ cells arise from local infoldings of surface germ cells (Seidel et al. 2018). Such infoldings can result in the local placement of more proximal germ cells adjacent to more distal germ cells. The PZ rachis appears to have a dense actin cytoskeleton, which may account for observed diffusion barriers (Cinquin et al. 2015; Gopal et al. 2017). Germ cells in the adult move from distal to proximal as a coherent (but not necessarily lineally related) group at a rate of ∼1 row/hr (Crittenden et al. 2006; Jaramillo-Lambert et al. 2007; Rosu and Cohen-Fix 2017) due to both displacement by distal germ cell divisions and continued removal of proximal oocytes by ovulation.
The hermaphrodite distal germ line is capped by a somatic cell called the DTC (Figure 1), which, in the adult, forms a “plexus” (an intricate network) where DTC cytoplasm and membranes intercalate between germ cells in the distal-most ∼4 cell diameter rows, followed by short intercalating processes up to ∼8 cell rows and long external processes that can extend ∼20 cell diameters (Byrd et al. 2014). Adult somatic sheath cells cover surface germ cells from leptotene through to the end of meiotic prophase, and appear to extend processes distally, partway into the PZ (Hall et al. 1999). The DTC, sheath cells, and germ cells are also covered by a basement membrane, providing integrity to the gonad. The DTC is the niche for the germline stem cells: the DTC expresses ligands for the GLP-1 Notch receptor that promotes the stem cell fate (see below). The DTC also provides small molecule factors for germ cell mitotic cycling/viability via gap junction channels formed by innexins expressed in the DTC and distal germ cells (Starich et al. 2014). E-cadherin and L1CAM adhesion proteins also localize to sites of DTC-germ cell contact, and each is required for the full complement of short intercalating processes (Gordon et al. 2019). The DTC thus polarizes the germ line, with the stem cells residing adjacent to the niche. The germ line also communicates with the DTC to ensure plexus formation and enwrapment of germ cells by the DTC (Linden et al. 2017).
Adult germline stem cell system: cell cycle properties
Cell transplantation and genetic lineage analysis have been important approaches for understanding cell fate and cellular dynamics in a number of stem cell systems. However, these approaches are not currently feasible for the C. elegans germ line. Instead, cellular behaviors have been inferred from cell population–based studies mostly in fixed preparations. These have employed measures of active M phase (e.g., nuclear morphology of mitotic figures or staining with anti-phospho-H3 antibodies) and S phase (e.g., the percentage labeling with cytologically detectable nucleotides such as EdU), or their indexes (the percentage of cells in active M or S phase). To provide dynamic information, these approaches have also been used in combination, in pulse, pulse-chase, and continuous EdU labeling experiments (Kocsisova et al. 2018). Groups of germ cells have been tracked in vivo using photo-convertible fluorescent proteins (Rosu and Cohen-Fix 2017), although single-cell tracking has not yet been achieved. These studies have provided information on the mitotic cell cycle and the cellular organization of the PZ.
PZ germ cells show continuous mitotic cell cycling (no quiescence), which is largely asynchronous (Crittenden et al. 2006; Maciejowski et al. 2006). The average mitotic index (MI) is not equal across the PZ: as a function of distance from the distal tip, the MI rises and falls over the first 10–12 cell diameters, and then falls further proximally. The average mitotic cell cycle is relatively short, ranging from 5 to 10 hr, with a median duration of ∼6–8 hr (Crittenden et al. 2006; Fox et al. 2011; Seidel and Kimble 2015; Rosu and Cohen-Fix 2017). Importantly, the proximal PZ contains noncycling cells in meiotic S phase (Crittenden et al. 2006; Jaramillo-Lambert et al. 2007; Fox et al. 2011); however, there are currently no markers that distinguish mitotic from meiotic S phase.
High mitotic cell cycle activity and a high rate of PZ cells entering the meiotic pathway are both required to generate a sufficient number of germ cells for peak progeny production in the young adult. The PZ is estimated to contain 130–160 mitotically cycling cells and 70–100 meiotic S phase cells, with ∼20 cells/hr entering meiotic prophase (PZ output) (Fox et al. 2011). A subset of meiotic prophase germ cells in the adult hermaphrodite function as nurse cells, providing cellular constituents to growing oocytes, and these undergo apoptosis in late pachytene (Gumienny et al. 1999; Wolke et al. 2007; Raiders et al. 2018). Nurse cells and oocyte precursor cells cannot be distinguished morphologically. Instead, their number can be estimated from PZ output and germline output (2.6 oocytes ovulated/hr/gonad arm), yielding ∼7 nurse cells per oocyte, with ∼85% meiotic prophase cells undergoing apoptosis (McCarter et al. 1999; Jaramillo-Lambert et al. 2007; Fox et al. 2011; Agarwal et al. 2018).
The germ cell cycle structure and regulation differs significantly from that of somatic cells. Germ cells have a very short or nonexistent G1 phase (Fox et al. 2011; Seidel and Kimble 2015), which has been confirmed by live imaging showing the appearance of DNA replication foci immediately after telophase (Lara-Gonzalez et al. 2019). In contrast, G1 is a prominent feature of the somatic cell cycle (Baugh and Sternberg 2006; Ruijtenberg and van den Heuvel 2015; van Rijnberk et al. 2017; Kipreos and van den Heuvel 2019). Germline stem cells in the Drosophila ovary similarly have a very short G1 (Hsu et al. 2008; Ables and Drummond-Barbosa 2013; Kao et al. 2015). Consistent with a short/absent G1, an important regulator of the G1-S transition CDK-4/Cyclin D is not required for germline proliferation in C. elegans (Fox et al. 2011). CDK-2/CYE-1 is required for germline mitotic cell cycling, but CYE-1 accumulation is not cell cycle–regulated, unlike in somatic cells where CYE-1 expression is limited to late G1 and S phase. Glycogen synthase kinase ortholog GSK-3 is required for high CDK-2 expression throughout the cell cycle, promoting rapid cell cycling (Furuta et al. 2018). Multiple environmental conditions that result in mitotic cell cycle quiescence all result in a G2 arrest (see below). Thus, the germ line appears to employ G2 as the phase for physiologically imposed cell cycle arrest, while somatic cells employ G1; the different cell cycle structure/regulation likely contributes to the distinct arrest points. Presumably, the G2 arrest promotes germline genome integrity, as sister chromatids can be used to repair DNA damage.
C. elegans has an asymmetric population-based germline stem cell system, contrasting with that of Drosophila
There are two general strategies stem cell systems utilize for self-renewal and the generation of differentiated cells, single-cell asymmetric cell division and population asymmetry, both of which can be governed by an extrinsic niche (Morrison and Kimble 2006; Simons and Clevers 2011). The well-characterized Drosophila male and female germ lines exhibit single-cell asymmetric stem cell divisions, where each stem cell division produces a niche-contacting cell that remains a stem cell and another cell that loses niche contact and will eventually differentiate. In contrast, the model most consistent with results for the C. elegans hermaphrodite germ line is population asymmetry, where strict lineal relationships between individual stem cells and their nonstem daughters is not required to maintain homeostasis. Rather, a pool of niche-governed stem cells proliferates and each stem cell division produces either two stem cells, one stem and one nonstem cell, or two nonstem cells. Because of the anatomical constraint of the distal gonad, proliferation causes proximal displacement of germ cells to a region outside the influence of the niche, and their subsequent differentiation. These different strategies are highlighted in Figure 3, showing schematic representations of germ cell behavior in the two systems (ignoring the complexities of the somatic cells in each system), and are described in more detail below.
In Drosophila a small number of germline stem cells (2–3 ovary; 6–12 testis) are physically attached to the niche (cap cells, ovary; hub cells, testis). The germline stem cells typically divide asymmetrically with respect to the niche such that the daughter that remains in contact with niche cells remains a stem cell, while the displaced nonstem daughter (cystoblast, ovary; gonialblast, testis) loses niche signaling and begins the path toward differentiation (Fuller and Spradling 2007; Losick et al. 2011; Laws and Drummond-Barbosa 2017). By contrast, the stem cell pool in the young adult C. elegans hermaphrodite is relatively large, estimated at 35–80 cells per gonad arm from population-based studies (Cinquin et al. 2010; Byrd et al. 2014; Fox and Schedl 2015) and 80–135 cells from GLP-1 Notch signaling transcriptional target proteins (see below; Shin et al. 2017; Kocsisova et al. 2019). Germ cell divisions appear to be symmetric with respect to fate. In addition, the division planes are not oriented with respect to the distal-proximal axis and cells throughout the PZ are equally sized (Crittenden et al. 2006). Electron microscopy has not revealed specialized attachments between distal germ cells and the DTC that could orient the cell division plane (Hall et al. 1999), although DTC-germ cell adhesion facilitates contact with niche projections (Gordon et al. 2019). The current model is that a large pool of stem cells divides symmetrically, and that the resulting divisions eventually displace germ cells proximally, away from contact with the DTC niche. This displacement results in loss of GLP-1 signaling activity and cells proceed toward meiotic entry.
The behavior of the nonstem cell daughters also differs between C. elegans and Drosophila (Figure 3). In Drosophila, the nonstem daughter of a stem cell division (the cystoblast or gonialblast) undergoes four stereotypical, transit-amplifying cell divisions, together as a cyst. The 16 progeny cells that are the products of these synchronous divisions then enter meiotic S phase and meiotic prophase (Fuller and Spradling 2007). In contrast, upon loss of DTC niche signaling, C. elegans nonstem cells appear to complete their ongoing mitotic cell cycle, enter meiotic S phase, and then meiotic prophase (Fox and Schedl 2015). This conclusion is based in part on the finding that following loss of GLP-1 signaling activity there is an approximate doubling of the number of mitotic cycling cells prior to meiotic entry. In addition, cells in mitotic S phase and G2 are not able to directly enter meiotic prophase without a mitotic division, presumably because of a requirement to reset the cell (minimally chromosomes and nuclear envelope) for meiotic events. Moreover, neither synchronous mitotic divisions nor cyst like structures are observed. Thus, in C. elegans there is essentially direct differentiation, likely requiring a single terminal division after loss of GLP-1 signaling activity.
Based on these results, the current model for the cellular organization of the stem cell system in the young adult hermaphrodite is that the distal germ line contains three pools of cells within the PZ, plus cells in meiotic prophase. The PZ contains a large stem cell pool that is actively responding to GLP-1, an equally sized pool of nonstem cells that have been displaced from DTC niche signaling and that are completing their final mitotic cell cycle, and a pool of cells undergoing meiotic S phase. The latter cells then progress to meiotic prophase (leptotene-zygotene) (Figure 2; Fox and Schedl 2015). These four pools of cells partially overlap in the adult, as indicated by overt meiotic entry into leptotene occurring over an ∼8 cell diameter “meiotic entry region” (Figure 2). The partial overlap is, at least in part, a consequence of stem cells being at different stages of the mitotic cycle when they reach a critical distance from the DTC niche. As a result, individual cells complete the ongoing mitotic cycle and begin meiotic S phase at different distal-proximal positions (Hansen et al. 2004a).
Stem Cell Fate vs. Meiotic Development Decision
We first briefly summarize the molecular network that governs germ cell identity within the PZ with respect to a cell that exhibits stem cell identity (or “stem cell fate”) vs. a cell that has entered the pathway toward meiotic development (Figure 4). After this summary, we then discuss the various regulatory steps in greater detail.
Figure 4.
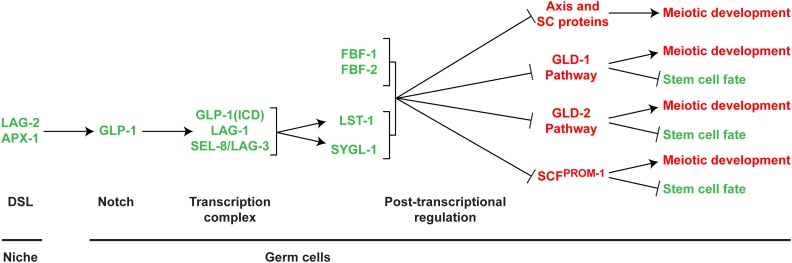
The genetic network for the stem cell fate vs. meiotic development decision. Top, model of the genetic network of gene products that promote the stem cell fate (green) by inhibition of three meiotic entry pathways, as well as inhibition of meiotic chromosome axis and SC protein expression (red), which together promote meiotic development. Arrows indicates positive regulation, lines with bar indicate inhibition. Bottom, regulatory class and location. DSL ligands, expressed in the niche/DTC, activate GLP-1 Notch in germ cells, leading to generation of GLP-1(ICD), which forms a transcriptional complex that results in spatially restricted expression of LST-1 and SYGL-1 that, together with ubiquitous PZ expressed FBF-1 and FBF-2, act in post-transcriptional repression of gene products that promote meiotic development.
DTC-germline signaling via the GLP-1 Notch pathway specifies the stem cell fate through repression of three redundant pathways that promote meiotic entry/development, the GLD-1 pathway, the GLD-2 pathway, and SCFPROM−1. Loss of DTC–GLP-1 Notch signaling results in all germline stem cells undergoing differentiation. Loss of any two of the three meiotic entry pathways (e.g., gld-1 gld-2 double mutant) results in ectopic proliferation or a tumor consisting of undifferentiated germ cells that have failed to enter meiotic prophase (see Appendix). The network exhibits extensive genetic redundancy, particularly downstream of canonical GLP-1 Notch signaling. This includes both nonparalogous redundancy (e.g., the meiotic entry pathways), and paralogous redundancy (e.g., Notch ligands). Similar to germ cell fate regulation in other organisms, the network involves many RNA-binding proteins and several different post-transcriptional regulation mechanisms. Some of these proteins and RNAs are found in granules, including P granules, which are germline-specific, membraneless RNA-protein condensates with liquid to hydrogel-like properties (Seydoux 2018). Therefore, a deeper understanding of the behavior of protein-RNA condensates and their components may further inform the network we describe below.
Regulatory Network: Stem Cell Fate
DTC niche: germline GLP-1 Notch signaling
In a classic developmental biology experiment, Judith Kimble and John White ablated the DTC using a laser microbeam to ask whether the somatic DTC signals the germ line (Kimble and White 1981). Killing the DTC caused all germ cells to enter meiotic prophase; that is, the stem cells were “lost” due to premature differentiation. Furthermore, repositioning or duplicating the DTC resulted in displaced or ectopic stem cells and a polarized assembly-line of meiotic development and gametogenesis (Kimble and White 1981; Kipreos et al. 2000; Lam et al. 2006). Thus, the DTC functions as the niche, being necessary and sufficient for both the stem cell fate and for the generation of a polarized germline differentiation assembly line.
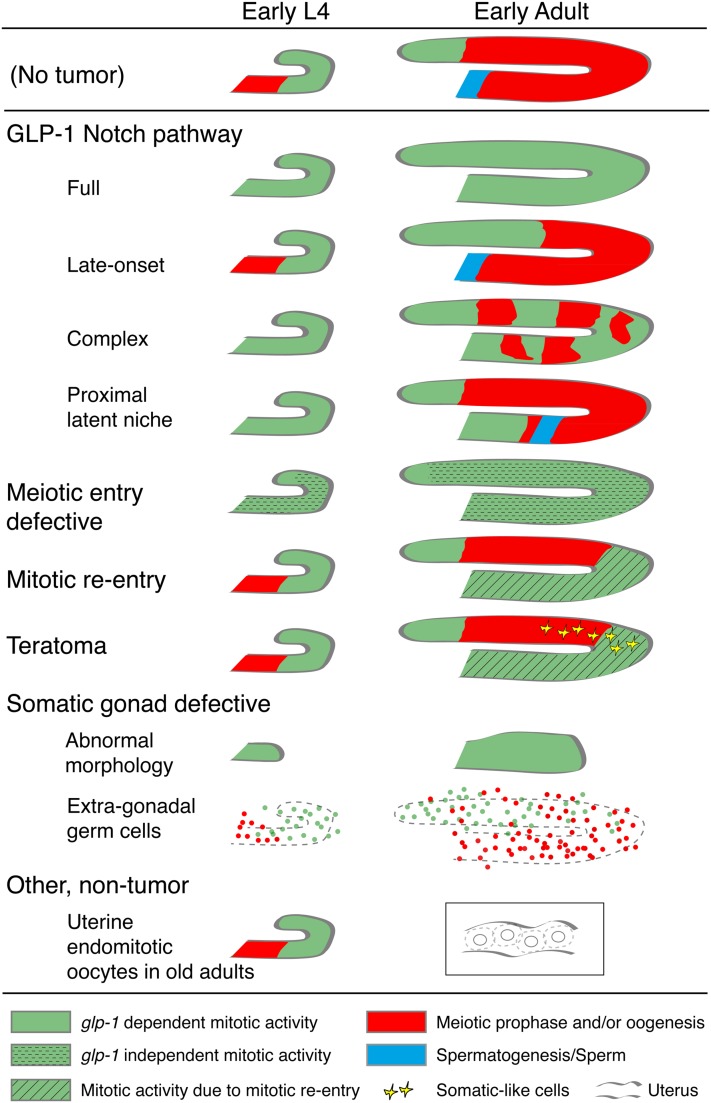
This “loss” of stem cells phenotype is called “Glp,” for Germline proliferation abnormal phenotype. However, this name is misleading as the phenotype was later understood to be a premature switch from the stem cell fate to meiotic development. The term “premature meiotic entry” can refer to temporally premature (that is, meiotic entry in early larvae prior to the normal time) or spatially premature (that is, meiotic entry at a position closer to the distal tip in later larvae and adults after establishment of the PZ; see below). The glp-1 mutant defect is not in proliferation or mitotic cell cycling. Defects in fate and cell cycle are genetically separable: germ cells can proliferate in the complete absence of glp-1, in combination with mutations in meiotic entry pathway genes (see below). Conversely, mutations in other glp genes (e.g., glp-3/eft-3/eef-1A.1, Kadyk et al. 1997; Maciejowski et al. 2005; glp-4, Beanan and Strome 1992; Rastogi et al. 2015) cause defects in proliferation but do not cause premature differentiation. Given these considerations, we suggest the phenotypic designation “Glp-1” to describe the “inappropriate switch of all stem cells from the stem cell fate to meiotic development,” distinct from a cell cycle progression phenotype that does not alter stem cell fate.
The DTC signals the germ line to specify the stem cell fate through the conserved Notch signaling pathway, via the Notch receptor homolog GLP-1 (Kimble and Crittenden 2007). GLP-1 is one of two Notch receptors in C. elegans (Greenwald and Kovall 2013). The other, LIN-12, functions redundantly with GLP-1 in some aspects of somatic development (Lambie and Kimble 1991; Fitzgerald et al. 1993; Priess 2005), but depletion of lin-12 by RNAi does not influence the size of the PZ (Pekar et al. 2017). Thus, GLP-1 appears to be the sole Notch receptor required for the germline stem cell fate. DTC ablation or loss of core GLP-1 Notch signaling components results in the Glp-1 phenotype. The identification of core Notch signaling genes in GLP-1–dependent stem cell fate was largely through screens for (i) mutants that displayed a Glp-1 phenotype singly or in combination with another GLP-1 pathway mutant, (ii) mutants that displayed other glp-1 phenotypes (e.g., embryonic anterior pharynx defect), (iii) mutants with a characteristic L1 arrest phenotype observed in the double loss of function (lf) of glp-1 and lin-12 (Lag, for lin-12 and glp-1), or (iv) mutants that are modifiers of various lin-12 phenotypes [reviewed in Greenwald 2012; Greenwald and Kovall 2013]. Below, we summarize germline GLP-1 signaling by the core Notch pathway genes (Table 1). In a number of cases, the molecular/biochemical activity of a given orthologous gene product has not been examined experimentally in C. elegans and its molecular function is assumed based on studies in mammalian systems and Drosophila (Kopan and Ilagan 2009).
Table 1. Genetics of the core stem cell fate vs. meiotic development decision gene network.
| Product/activitya | Human homolog | Reference(s) | |
|---|---|---|---|
| Genes that promote the stem cell fate | |||
| Canonical Notch signalingb | |||
| Receptor | |||
| glp-1 | Notch receptor | Notch 1-4 | Austin and Kimble (1987); Yochem and Greenwald (1989) |
| Ligands | |||
| lag-2 | DSL-like ligand | DSL | Lambie and Kimble (1991); Tax et al. (1994) |
| apx-1 | DSL-like ligand | DSL | Mango et al. (1994); Nadarajan et al. (2009) |
| Ligand activity | |||
| epn-1 | Ligand endocytosis | EPN1-2 | Tian et al. (2004) |
| mib-1 | Ubiquitin ligase homolog | MIB1 | Ratliff et al. (2018) |
| Site 2 protease | |||
| sup-17 | Disintegrin and metalloproteinase domain | ADAM10 | Tax et al. (1997); Wen et al. (1997) |
| adm-4 | Disintegrin and metalloproteinase domain | ADAM17 | Jarriault and Greenwald (2005) |
| Site 3 protease | |||
| sel-12 | Presenilin | PSEN2 | Levitan and Greenwald (1995) |
| hop-1 | Presenilin | PSEN2 | Li and Greenwald (1997); Westlund et al. (1999) |
| aph-1 | Gamma-secretase subunit | APH1A | Goutte et al. (2002); Francis et al. (2002) |
| aph-2 | Nicastrin | NCSTN | Goutte et al. (2000); Levitan et al. (2001) |
| pen-2 | Gamma-secretase subunit | PSENEN | Francis et al. (2002) |
| Nuclear complex | |||
| lag-1 | DNA-binding protein | CBF1 | Lambie and Kimble (1991); Christensen et al. (1996) |
| sel-8 (aka lag-3) | Mastermind-like transcriptional coactivator | MAML1 | Tax et al. (1997); Doyle et al. (2000); Petcherski and Kimble (2000) |
| Downstream/in parallel of GLP-1 signaling | |||
| lst-1c | Novel | — | Kershner et al. (2014); Lee et al. (2016) |
| sygl-1c | Novel | — | Kershner et al. (2014); Lee et al. (2016) |
| fbf-1d | Puf RNA-binding domain | — | Zhang et al. (1997); Crittenden et al. (2002); Bernstein et al. (2005) |
| fbf-2d | Puf RNA-binding domain | — | Zhang et al. (1997); Crittenden et al. (2002); Bernstein et al. (2005) |
| Genes that promote meiotic entrye | |||
| GLD-1 pathway | |||
| gld-1 | Maxi-KH domain RNA-binding protein | QKI | Francis et al. (1995a,b) |
| nos-3 | Nanos RNA-binding protein | — | Kraemer et al. (1999); Hansen et al. (2004b) |
| GLD-2 pathway | |||
| gld-2 | GLD-2 family cytoplasmic polyA polymerase | TENT2 | Kadyk and Kimble (1998); Wang et al. (2002) |
| gld-3 | KD domain RNA-binding protein | — | Eckmann et al. (2002), (2004); Suh et al. (2006) |
| SCFPROM−1 pathway | |||
| prom-1 | F-box protein | FBXO47 | Jantsch et al. (2007); Mohammad et al. (2018) |
| skr-1/skr-2 | SKP1 SCF complex subunit | SKP1 | Nayak et al. (2002); Mohammad et al. (2018) |
| cul-1 | Cullin SCF complex subunit | CUL1 | Kipreos et al. (1996); Mohammad et al. (2018) |
Genotype and phenotype that supports the indicated function in the network is described in the corresponding footnotes.
Single gene null (e.g., glp-1), partial loss of function (e.g., lag-2), or RNAi (e.g., lag-1), or in combination with a ts glp-1 allele at the permissive temperature (noted in the text as “glp-1(rf-ts) pt”), results in elevated penetrance of the Glp-1 meiotic entry phenotype in which all stem cells differentiate. This enhancement, while supporting that the gene functions in Notch signaling to promote the stem cell fate, does not demonstrate that the gene is required for signaling activity in this context.
The lst-1 sygl-1 null double mutant results in a Glp-1 premature meiotic entry phenotype.
The fbf-1 fbf-2 null double mutant results in all germ cells entering meiosis in the late L4 stage, at 20°.
Genetic loss of genes in any two pathways (e.g., gld-1; gld-3 or gld-1 prom-1) results in a tumorous phenotype due to a failure of germ cells to enter meiosis.
The DTC expresses two Notch ligands, the single-pass transmembrane DSL (for Delta/Serrate/LAG-2) proteins LAG-2 and APX-1 (Henderson et al. 1994; Tax et al. 1994; Nadarajan et al. 2009). Genetic and expression studies indicate that DTC-expressed LAG-2 activates germline GLP-1 in early larvae and that APX-1 and LAG-2 redundantly activate GLP-1 in later larval and adult life. LAG-2 is found in puncta on adult DTC membrane and processes, consistent with the large stem cell pool controlled by the DTC niche (Gordon et al. 2019). LAG-2 and APX-1 contain two or three EGF repeats, respectively, and a single DSL domain, similar to DSL family ligands in other animals. However, LAG-2 and APX-1, as well as other C. elegans DSL ligands, lack the DOS motif (for Delta and OSM-11) found in vertebrate and Drosophila ligands (Chen and Greenwald 2004; Komatsu et al. 2008). Instead, C. elegans has single-pass transmembrane or secreted proteins that contain the DOS domain, at least one of which, OSM-11, functions in LIN-12 Notch signaling (Komatsu et al. 2008). The reason C. elegans has bipartite Notch ligands is not known. The role of the five identified DOS domain–containing proteins in DTC-germline signaling to promote the stem cell fate remains to be investigated.
The GLP-1 receptor is on the surface of germ cells throughout the PZ, as well as on internal subcellular vesicular compartments (Crittenden et al. 1994). However, as described below, active GLP-1 signaling (transcription of target genes) is more spatially restricted, limited to the distal-most ∼5 cell diameters where there is intimate DTC contact and thus high ligand level. GLP-1 function is required continuously for the stem cell fate: in glp-1 null mutant hermaphrodites and males, all germ cells enter meiosis prematurely in early larvae, both temporally premature (with respect to the normal time of meiotic entry during larval development) and spatially premature (with respect to the DTC) entry of all stem cells into the meiotic pathway. A shift of glp-1 reduction-of-function (rf) temperature-sensitive (ts) mutants to the restrictive temperature at any time during larval or adult life results in all the stem cells entering meiosis (Austin and Kimble 1987; Kodoyianni et al. 1992). Such glp-1(rf-ts) mutants at a permissive temperature (pt; this combination hereafter abbreviated as “glp-1(rf-ts) pt”) are fertile and have a smaller PZ, consistent with a reduced stem cell pool (due to meiotic entry at a spatial position closer to the distal tip), but have wild-type mitotic germ cell cycle kinetics (Michaelson et al. 2010; Fox and Schedl 2015; Lee et al. 2016). Thus, while GLP-1 signaling functions in specification of the stem cell fate, it apparently has no role in controlling the rate of mitotic cell cycle progression.
When DTC-expressed LAG-2 or APX-1 interact with GLP-1 on germ cells, ligand-dependent cleavage of the receptor results in the release of the GLP-1 intracellular domain, called GLP-1(ICD), which then translocates to the nucleus. Signaling mediated proteolytic cleavage of the GLP-1 receptor occurs at two positions, site 2 and 3 (Greenwald and Kovall 2013). Site 2 is in the extracellular domain adjacent to the transmembrane domain, with cleavage catalyzed by A Disintegrin And Metalloprotease domain (ADAM) protease family members SUP-17, which is orthologous to Drosophila Kuzbanian and mammalian ADAM10, and by ADAM-4, which is orthologous to mammalian TACE/ADAM17 (Tax et al. 1997; Wen et al. 1997; Jarriault and Greenwald 2005). SUP-17 and ADAM-4 appear to function redundantly in germline GLP-1 signaling. Following ligand-dependent site 2 cleavage, γ-secretase performs cleavage at site 3 in the transmembrane domain. C. elegans γ-secretase is composed of catalytic subunit paralogs SEL-12 and HOP-1, which are orthologous to mammalian Presenilin 1 and 2, and associated subunits APH-1, APH-2, and PEN-2, which are orthologous to mammalian Aph1, Nicastrin, and Pen2, respectively (Levitan and Greenwald 1995; Li and Greenwald 1997; Westlund et al. 1999; Goutte et al. 2000, 2002; Levitan et al. 2001; Francis et al. 2002). SEL-12 and HOP-1 act redundantly in GLP-1 signaling during larval development, with HOP-1 functioning predominantly in adulthood (Agarwal et al. 2018). Additionally, EPN-1, the Epsin endocytosis protein homolog, and MIB-1, the Mind Bomb E3 ubiquitin ligase homolog, apparently function in the DTC/signaling cell to promote efficient ligand-dependent signaling, based on work in other systems (Tian et al. 2004; Kovall et al. 2017; Ratliff et al. 2018).
Following ligand-dependent cleavage, GLP-1(ICD) translocates into the nucleus and associates with the sequence-specific DNA-binding protein LAG-1 (Lambie and Kimble 1991; Christensen et al. 1996), a founding member of the CSL family of proteins [for CBF1 (also called RBPJ) in mammals, Su(H) in Drosophila, and LAG-1 in worms]. The LAG-1 DNA-binding sequence motif is essentially the same as that found for orthologous CSL proteins (Christensen et al. 1996). GLP-1(ICD) and LAG-1 bind in a ternary complex with SEL-8 (also called LAG-3), which may have the same function as Drosophila and mammalian Mastermind, forming a transcriptional activation complex (Tax et al. 1997; Doyle et al. 2000; Petcherski and Kimble 2000). In other systems, CSL can also function as a transcriptional repressor in the absence of Notch activation; it is not known if LAG-1 functions in transcriptional repression in C. elegans.
The behavior of the glp-1 gain-of-function (gf) allele oz112 supports the model for germline Notch signaling described above (Berry et al. 1997). glp-1(oz112gf) mutants have an overproliferation or tumorous germline phenotype (see Appendix) that arises from germ cells failing to enter meiosis, the phenotypic opposite of inappropriate meiotic entry observed with glp-1 lf, indicating that GLP-1 activity is necessary and sufficient to promote the stem cell fate/inhibit meiotic development. A number of lines of evidence demonstrate that the GLP-1(oz112gf) Ser642Asn missense mutant protein is constitutively active, signaling independent of ligand, including (i) retention of the tumorous germline phenotype following loss of the DTC or lag-2 (Berry et al. 1997); (ii) the position of the mutation in the extracellular N-terminal heterodimerization domain, a part of the negative regulatory region (NRR) that keeps the receptor inactive in the absence of ligand (Greenwald and Kovall 2013); and (iii) the ability of the homologous mammalian Notch1 mutant oz112 to undergo ligand-independent S2 cleavage in HEK293 cells (Vooijs et al. 2004; van Tetering et al. 2009). Notably, similar activating missense mutations in the NRR region of Notch1 result in human T cell acute lymphoblastic leukemia (Weng et al. 2004). glp-1(oz112gf) homozygote germ lines contain only undifferentiated mitotically cycling stem cells, in larvae and adults and in hermaphrodites and males (Berry et al. 1997; Hansen et al. 2004b; Lee et al. 2016). By contrast, in many glp-1(oz112gf) heterozygotes or hemizygotes, germ cells enter meiosis normally during larval development, but the stem cell pool expands with time beyond 20 cell diameters PZ by the young adult stage. This phenotype is called a late-onset tumorous phenotype (Berry et al. 1997; Pepper et al. 2003a; Hansen et al. 2004b). See Appendix for a description of the various types of germline overproliferation/tumorous phenotypes. Because of the strongly reduced fertility in glp-1(oz112gf) heterozygotes and hemizygotes, this allele is difficult to work with.
There is a group of ts glp-1(gf-ts) mutants that are more experimentally tractable, with ar202 being the most widely used allele (Pepper et al. 2003a,b; Kerins et al. 2010). They also contain missense changes in the NRR and thus are predicted to undergo ligand independent signaling. While genetic evidence supports a degree of ligand-independent cleavage (Pepper et al. 2003a), glp-1(ar202) is also responsive to ligand (McGovern et al. 2009). The allele displays several unusual genetic characteristics for a gf allele, including a recessive character and both maternal and zygotic dosage and temperature effects (Pepper et al. 2003a). A highly penetrant phenotype observed in glp-1(ar202gf) at elevated temperatures is overproliferation in the proximal part of the germ line (Pro phenotype), together with the normal pattern of germline development, including spermatogenesis and then oogenesis (see Appendix). The glp-1(ar202gf) Pro phenotype is the result of proximal germ cells failing to enter meiotic prophase in the L3 stage (Pepper et al. 2003b) (see Appendix, Latent niche). However, depending on rearing conditions, glp-1(ar202) can display the late-onset tumorous phenotype with or without the Pro phenotype. glp-1 ar202gf differs from oz112gf in a number of ways: ar202 mutants show less overproliferation/more meiotic entry than oz112gf mutants, the ar202gf overproliferation phenotype is suppressed by a wild-type allele while oz112gf is enhanced, and the ar202gf germline remains responsive to ligand (Berry et al. 1997; Pepper et al. 2003a,b; Hansen et al. 2004a).
GLP-1 Notch transcriptional targets lst-1 and sygl-1
Two germline transcriptional targets of GLP-1 signaling have been identified, lst-1 and sygl-1 (Kershner et al. 2014; Lee et al. 2016). lst-1 is also a LIN-12 transcriptional target in vulval development (Yoo et al. 2004). LST-1 and SYGL-1 are novel proteins limited to the Caenorhabditis clade. LST-1 also contains a single Nanos-like zinc finger, suggesting that it may function in post-transcriptional regulation, like other Nanos proteins (Kershner et al. 2014). SYGL-1 and LST-1 are redundantly necessary and each is sufficient for promoting the stem cell fate. lst-1 and sygl-1 null single mutants are phenotypically largely wild type, while the double mutant displays the Glp-1 early larval premature meiotic entry phenotype identical to the glp-1 null. In contrast, ubiquitous germline overexpression of either LST-1 or SYGL-1 results in an overproliferation phenotype (Shin et al. 2017). Genetically, LST-1 and SYGL-1 act downstream of GLP-1 [see Kershner et al. (2014); Shin et al. 2017 for details]. Molecularly, lst-1 and sygl-1 transcription is restricted to the distal-most region of the germ line and requires GLP-1 activity (see below; Kershner et al. 2014; Lee et al. 2016). Furthermore, lst-1 and sygl-1 loci contain multiple LAG-1 DNA-binding sites. For sygl-1, these sites were tested and found to be required for distal germline expression in a transgene assay (Kershner et al. 2014). It is not known if there exist additional germline transcriptional targets of GLP-1 signaling.
An important issue that was resolved once GLP-1 Notch pathway transcriptional targets were in hand is which cells in the PZ are actively responding to GLP-1 signaling. Lee et al. (2016) employed single-molecule fluorescent in situ hybridization using intron probes to identify cells that contain nuclear foci representing nascent sygl-1 and lst-1 transcripts and to quantify foci intensity. In the first three cell diameters from the distal tip, ∼70% of nuclei have between 1 and 4 nascent transcript foci, followed by a steep fall to background levels by 6–7 cell diameters (Figure 2). The few nuclei in this region that do not have nascent transcript foci are in cells that nevertheless have cytoplasmic sygl-1 and lst-1 messenger RNA (mRNA). These results suggest that GLP-1 signaling occurs through a transcriptional pulsing mechanism, where transcription occurs episodically, even for active genes (Elowitz et al. 2002; Raj et al. 2006). Such a mechanism had not been reported previously for Notch-mediated transcriptional activation. In addition, glp-1(oz112gf) tumorous germ lines display a constant number and intensity of sygl-1 foci throughout the germ line, similar to that observed in the distal-most three cell diameters in the wild type, consistent with a loss of spatial regulation rather than an increase in transcriptional output. In wild type, intimate DTC contact with germ cells is highly correlated with GLP-1 transcriptional output, within ∼5 cell diameters from the distal tip (Lee et al. 2016).
LST-1 and SYGL-1 are cytoplasmic proteins. LST-1 is found in the first five cell diameters from the distal tip, at a relatively uniform level, and its distribution is similar to cytoplasmic lst-1 mRNA (Shin et al. 2017). SYGL-1 distribution is somewhat broader; it is high in the first ∼5 cell diameters and then falls off more gradually to 10–13 cell diameters (Figure 5; Shin et al. 2017; Kocsisova et al. 2019). Consistent with SYGL-1 and LST-1 each being sufficient for the stem cell fate and consistent with their differences in expression pattern, the PZ of the lst-1 null single mutant is normal, likely due to the presence of SYGL-1, while the PZ of the sygl-1 null single mutant is shorter, likely due to the more spatially restricted expression pattern of remaining LST-1. While the onset of sygl-1 expression is regulated transcriptionally by GLP-1 signaling, the level and extent of SYGL-1 protein accumulation is limited post-transcriptionally, at least in part by its 3′UTR (Shin et al. 2017). The gene products that function in sygl-1 3′UTR regulation are not known. The proximal extent of SYGL-1 and LST-1 is also limited by protein degradation, although the genes involved are not known.
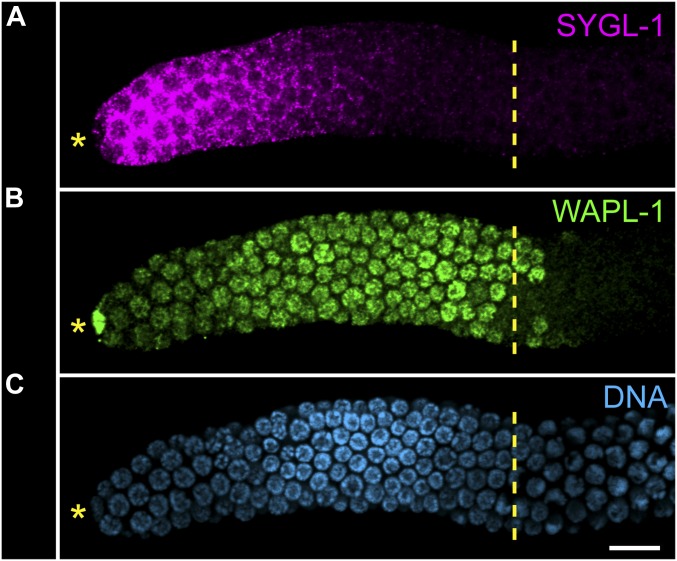
Figure 5.
SYGL-1 accumulation in the young adult progenitor zone. Spatially restricted, but nonuniform accumulation of SYGL-1 in the PZ of a dissected young adult hermaphrodite gonad. (A) Cytoplasmic SYGL-1 (pink, SYGL-1::3×FLAG); (B) PZ cell nuclei, as well as the DTC nucleus (green, WAPL-1 staining); (C) all cell nuclei (blue, DAPI). Yellow dashed vertical line, boundary of the PZ and leptotene. Bar, 10 µM. Figure from Zuzana Kocsisova.
FBF
Two PUF (for Pumilio and FBF) family mRNA-binding protein paralogs, FBF-1 and FBF-2, promote the germline stem cell fate (in most cases they will be referred to jointly as FBF) (Kershner et al. 2013). FBF-1 and FBF-2 are cytoplasmic proteins that function throughout the PZ and into meiotic prophase (Crittenden et al. 2002, 2017; Suh et al. 2009; Merritt and Seydoux 2010; Arur et al. 2011; Voronina et al. 2012). By comparison with the spatially restricted GLP-1 transcriptional targets LST-1 and SYGL-1, FBFs are detected throughout the PZ (Figure 2 and Figure 6). FBF-1 and FBF-2 are redundantly necessary to promote the stem cell fate in late larvae and adults; although null single mutants are largely wild type, fbf-1 fbf-2 null double mutants display a Glp-1–like meiotic entry phenotype starting in the late L4 stage (Crittenden et al. 2002; Lamont et al. 2004). With some exceptions (see below), FBF-1 and FBF-2 display similar biochemical functions, binding to the same RNA motif (the FBE, for FBF-binding element), and binding to the GLD-3 protein (Zhang et al. 1997; Eckmann et al. 2004; Bernstein et al. 2005).
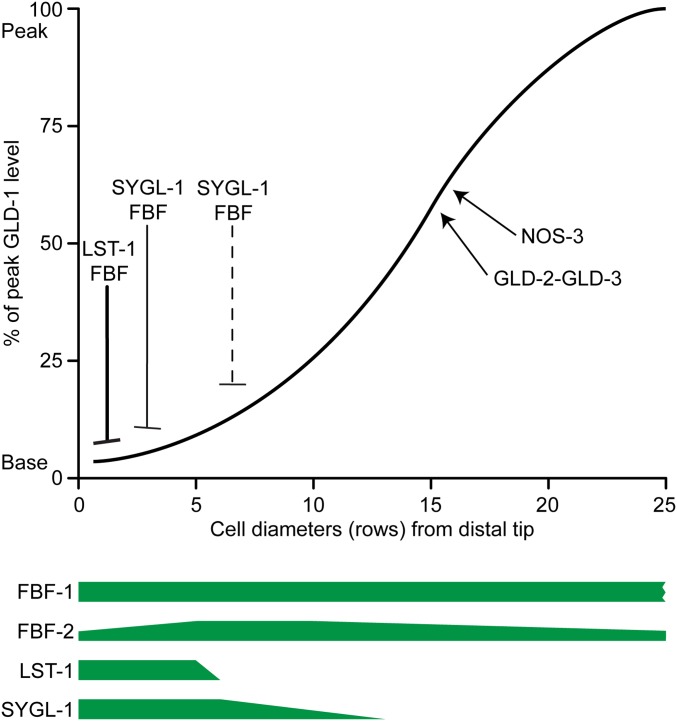
Figure 6.
Control of GLD-1 accumulation in the progenitor zone. Model describing spatial control of GLD-1 accumulation in the young adult hermaphrodite. Spatial pattern of repressors of GLD-1 accumulation in the PZ: LST-1 and SYGL-1 accumulation are spatially restricted, while FBF-1 and FBF-2 accumulate throughout the PZ. LST-1 and SYGL-1 are proposed to limit the activity of FBF-mediated repression of GLD-1 to the distal-most region of the PZ. Activators of GLD-1 accumulation, NOS-3 and GLD-2/GLD-3, function redundantly with each other and accumulate essentially throughout the PZ (not shown). See text for details.
More than 1200 mRNAs have been identified that bind to FBF in genome-wide RNA pulldown analysis (Kershner and Kimble 2010). Most of these mRNA targets do not have a known function in the stem cell vs. meiotic development decision, and at least some are regulated by FBF to control other aspects of germline biology, including the fem-3 mRNA in germline sex determination and membrane organization during oogenesis (Zhang et al. 1997; Arur et al. 2011). Importantly, two sets of targets are regulated by FBF for the stem cell vs. meiotic development switch (Figure 4). First, FBF regulates meiotic chromosome structural protein mRNAs that function in early steps of meiotic development [e.g., chromosome axis protein HIM-3, and synaptonemal complex (SC) proteins SYP-2 and SYP-3; Merritt et al. 2008; Kershner and Kimble 2010; Merritt and Seydoux 2010]. FBF post-transcriptionally represses the accumulation of these proteins in the distal PZ. Hence, they begin accumulation in the proximal PZ and then commence loading onto chromosomes at overt meiotic entry in leptotene (Merritt and Seydoux 2010). Second, FBF regulates mRNAs of genes that function in the meiotic entry pathway, including gld-1, gld-2, and gld-3, which are repressed in the distal PZ to inhibit meiotic development.
The best-characterized FBF mRNA target is gld-1. FBF activity inhibits distal GLD-1 accumulation and therefore prevents distal meiotic entry. Upon loss of FBF or loss of FBEs in the gld-1 3′UTR, high GLD-1 is observed throughout the PZ and premature meiotic entry occurs (Crittenden et al. 2002; Hansen et al. 2004b; Suh et al. 2009; Brenner and Schedl 2016). This distal repression of GLD-1 by FBF is recapitulated with reporter transgenes containing the gld-1 3′UTR in an FBE-dependent manner (Merritt et al. 2008; Merritt and Seydoux 2010). Molecularly, FBF binds the gld-1 3′UTR in vitro and gld-1 mRNA in vivo (Crittenden et al. 2002; Suh et al. 2009). Similar to other PUF family proteins, FBF recruits the Ccr4/Not deadenylase complex to destabilize the gld-1 mRNA (Suh et al. 2009).
However, notwithstanding the 89% overall sequence identity and redundant genetic functions, FBF-1 and FBF-2 have distinct roles in gld-1 regulation and distinct subcellular localization. FBF-1 appears to specifically function in gld-1 mRNA destabilization since the fbf-1 null single mutant, but not the fbf-2 single null mutant, has elevated gld-1 mRNA in the distal germ line (Voronina et al. 2012). FBF-2 appears to specifically function in translational repression in conjunction with the P granule component PGL-1 (Voronina et al. 2012). With respect to localization, FBF-1 is found in numerous cytoplasmic as well as perinuclear foci, which are distinct from P granules, whereas FBF-2 is primarily perinuclear and colocalizes with PGL-1/P granules (Voronina et al. 2012). Thus, differences in FBF-1 and FBF-2 functions are likely due to different binding partners and complexes in which they assemble.
A number of lines of evidence indicate that SYGL-1 and LST-1 function with FBF to promote the stem cell fate/repress meiotic development via GLD-1 (Figure 4). SYGL-1 and LST-1 function depends on FBF: the overproliferation observed from ectopic expression of SYGL-1 or LST-1 is suppressed by FBF loss of function, and FBF-1 and FBF-2 bind SYGL-1 and LST-1 (Shin et al. 2017). Importantly, SYGL-1 and LST-1 function in post-transcriptional repression of GLD-1, as evidenced by (i) pulldown of gld-1 mRNA and (ii) elevated distal GLD-1 levels in sygl-1 and lst-1 null single mutants, with peak levels throughout the distal 20 cell diameters in double mutants (Brenner and Schedl 2016; Shin et al. 2017). This leads to the model, elaborated below, that SYGL-1 and LST-1 spatially limit ubiquitous FBF post-transcriptional repression activity to the distal-most germ cells. Presumably, SYGL-1 and LST-1 also collaborate with FBF in post-transcriptional repression of meiotic chromosome axis and SC protein mRNAs.
Regulatory Network: Meiotic Development
Three post-transcriptional pathways, the GLD-1, GLD-2, and SCFPROM−1 pathways, act in concert to promote meiotic development (Figure 4 and Table 1). They function redundantly: following genetic loss of any two pathways, germ cells fail to enter meiotic prophase and undifferentiated cells proliferate ectopically and can form a “meiotic entry defective” type of germline tumor (see Appendix). In contrast, germ cells enter meiotic prophase more or less normally following loss of any single pathway gene member. The three pathways are placed downstream of and are inhibited by GLP-1 signaling. This conclusion is based on the observation that the overproliferation phenotype caused by loss of any two meiotic development pathway genes is epistatic to the glp-1 null or lst-1 sygl-1 null double, with respect to Glp-1–like meiotic entry phenotypes. Thus, DTC–GLP-1 signaling promotes the stem cell fate by inhibiting meiotic development.
GLD-1 pathway
GLD-1 is a KH RNA-binding domain containing protein that is homologous to the mammalian RNA-binding protein Quaking (Jones and Schedl 1995; Lee and Schedl 2010). gld-1 null single mutant hermaphrodites, but not males, have a tumorous germ line of the “mitotic reentry” type (see Appendix). The hermaphrodite-specific tumorous germ line was initially confusing until it was determined that during oogenesis, germ cells entered meiosis normally but a tumor arose from pachytene cells exiting meiotic prophase and reentering the mitotic cell cycle (Ellis and Kimble 1995; Francis et al. 1995a,b; Jones et al. 1996). In contrast, a gld-1 null in combination with single mutants in the GLD-2 or SCFPROM−1 pathways results in tumorous phenotypes of the “meiotic entry defective” class (see Appendix). These latter tumors are independent of sexual identity of the germline or soma (Kadyk and Kimble 1998; Hansen et al. 2004a; Mohammad et al. 2018). Such analyses illustrate the importance of determining the cellular origins of overproliferation phenotypes for correct interpretation of gene function (see Appendix). Normally, GLD-1 is at a low basal level in distal-most germ cells, rising ∼20-fold in a sigmoidal pattern and peaking just after overt meiotic entry in leptotene at 23–28 cell diameters from the distal tip (Figure 6) (Jones et al. 1996; Brenner and Schedl 2016). While GLD-1 is redundantly necessary to promote meiotic development, high GLD-1 levels in distal-most germ cells is sufficient to force stem cells to enter meiosis (Crittenden et al. 2002; Hansen et al. 2004b; Brenner and Schedl 2016; Shin et al. 2017).
GLD-1 is cytoplasmically localized and functions in translational repression, largely but not exclusively through 3′UTR regulation (Jan et al. 1999; Lee and Schedl 2001, 2004; Marin and Evans 2003; Biedermann et al. 2009; Jungkamp et al. 2011; Wright et al. 2011; Theil et al. 2018). More than 1000 GLD-1 mRNA targets have been identified, but it is unclear which are repressed to promote meiotic entry. For example, GLD-1 represses glp-1 mRNA translation; however, this downregulation occurs proximal to the PZ, after the switch to meiotic entry has occurred. GLD-1 also represses cye-1 mRNA translation, but only after meiotic entry in late zygotene and pachytene.
NOS-3, which is an ortholog of the Drosophila and mammalian RNA-binding protein Nanos (Kraemer et al. 1999), is placed in the GLD-1 pathway based on the observation that loss of nos-3 behaves similarly to the gld-1 null in double mutants with GLD-2 and SCFPROM−1 pathway genes (Hansen et al. 2004b; Mohammad et al. 2018). NOS-3 is cytoplasmic, and expressed throughout the germ line. Although NOS-3 functions with FBF in translational repression of the fem-3 RNA in germline sex determination and oocyte membrane organization (Zhang et al. 1997; Arur et al. 2011), NOS-3 and FBF act antagonistically in the stem cell vs. meiotic development decision, as the nos-3 null strongly suppresses the Glp-1–like meiotic entry phenotype of the fbf-1/-2 null double mutant (Eckmann et al. 2004; Hansen et al. 2004b). NOS-3 promotes peak GLD-1 accumulation in the proximal PZ, through a process that is redundant with the GLD-2 pathway (Hansen et al. 2004b; Brenner and Schedl 2016). How NOS-3 promotes GLD-1 accumulation, and what other targets it regulates to promote meiotic entry are unknown.
GLD-2 pathway
GLD-2 is a cytoplasmic polyA polymerase (Kadyk and Kimble 1998; Wang et al. 2002; Nousch et al. 2017). Also in the GLD-2 pathway is GLD-3, a Bicaudal-C family RNA-binding protein (Eckmann et al. 2002, 2004; Suh et al. 2006). GLD-3 associates with GLD-2, promotes binding to specific mRNA substrates (e.g., gld-1), and stimulates basal GLD-2 polyA polymerase activity (Suh et al. 2006; Schmid et al. 2009). GLD-2 and GLD-3 are positive regulators of peak GLD-1 accumulation in the proximal PZ (Figure 6) (Hansen et al. 2004b; Suh et al. 2006, 2009; Brenner and Schedl 2016). In addition to promoting gld-1 mRNA translation through polyA elongation, GLD-2 and GLD-3 appear to elevate gld-1 mRNA levels in a process that opposes CCR4/Not mediated mRNA degradation (see above; Suh et al. 2006; Millonigg et al. 2014; Nousch et al. 2014). However, the GLD-2 pathway must have other functions to promote meiotic entry because the gld-1 and gld-2 single null mutants are proficient in meiotic entry while the double mutant is not. More than 500 GLD-2 mRNA targets have been identified, but it is not known which targets besides gld-1 function in meiotic entry (Kim et al. 2010).
GLD-4, aTrf4/5-type cytoplasmic polyA polymerase, and GLS-1, a novel protein, function as a subcomponent of the GLD-2 pathway, apparently acting genetically redundantly with GLD-2, promoting peak GLD-1 accumulation through polyA-stimulated translation and stabilization of the gld-1 mRNA (Schmid et al. 2009; Rybarska et al. 2009; Millonigg et al. 2014). However, because gld-4 and gls-1 null mutants do not show a synthetic tumorous/meiotic entry defect with gld-1 or nos-3 null mutants, unlike gld-2 and gld-3, they are considered as a subcomponent of the GLD-2 pathway activity. GLD-4 also promotes the stem cell fate as a positive regulator of glp-1 mRNA translation (Millonigg et al. 2014).
SCFPROM−1 pathway
SCFPROM−1 is an E3 ubiquitin-ligase complex containing the Skp homologs SKR-1/2 (Nayak et al. 2002); CUL-1, the founding member of the Cullin family (Kipreos et al. 1996); and the F-box protein PROM-1 (Jantsch et al. 2007), which directs meiotic development through regulation of two biological processes (Mohammad et al. 2018). First, SCFPROM−1 downregulates mitotic cell cycle proteins, including CYE-1, WAPL-1, KNL-2, and pCDC-6, at the boundary of the PZ and leptotene. CYE-1 appears to be a direct substrate for ubiquitin mediated degradation by SCFPROM−1; it is not known if the other proteins are directly or indirectly regulated. In the absence of SCFPROM−1, these mitotic cell cycle proteins fail to be downregulated at meiotic entry, and exhibit an expanded expression pattern to more than 15 cell diameters proximal to the normal boundary. Remarkably, notwithstanding the ectopic accumulation of mitotic cell cycle proteins upon loss of SCFPROM−1 function, germ cells do not cycle mitotically. However, in double mutants of SCFPROM−1 with null alleles in either GLD-1 pathway or GLD-2 pathways genes, ectopic proliferation occurs and is epistatic to the premature meiotic entry phenotype of the glp-1 null. Second, SCFPROM−1 promotes pairing of homologous chromosomes at meiotic entry. CHK-2 is a master regulator of pairing in C. elegans (MacQueen and Villeneuve 2001; Kim et al. 2015). In the absence of SCFPROM−1, multiple CHK-2-dependent activities required for pairing fail to occur, while other activities associated with meiotic entry do occur (e.g., expression of meiotic chromosome axis protein HIM-3; upregulation of GLD-1 accumulation). SCFPROM−1 appears to promote homolog pairing as an indirect positive regulator of CHK-2 (V. Jantsch, Y. Kim, personal communication). Genetic analysis indicates that downregulation of mitotic gene products and promotion of pairing are separately controlled by SCFPROM−1, likely through different substrates. It is not currently known how GLP-1 signaling controls SCFPROM−1 activity.
The emerging picture is that meiotic entry occurs through a redundant post-transcriptional network, which coordinately regulates many gene products to achieve this complex differentiation process. There are two areas where there are significant gaps in our knowledge. First, while there is evidence of cross-regulation, by which the GLD-2 pathway functions in parallel with NOS-3 to promote GLD-1 accumulation, there is likely additional cross regulation that coordinates different processes. For example, the GLD-1 and GLD-2 pathways may regulate SCFPROM−1 activity. Second, the targets of the GLD-1 and the GLD-2 pathways that promote meiotic entry remain to be identified. While some SCFPROM−1 direct and indirect substrates have been identified, there are certainly more.
Regulatory Network: The Switch
A current model for control of stem cell fate and the switch to meiotic development is shown in Figure 4 and Figure 6. The DTC promotes the stem cell fate by spatially restricting activation of ligand-dependent cleavage of GLP-1, generating GLP-1(ICD), which acts together with LAG-1 and SEL-8 to transcribe lst-1 and sygl-1 in the distal-most ∼5 cell diameters. SYGL-1 accumulates more broadly than LST-1, extending as far as the distal-most 10–13 cell diameters. SYGL-1 and LST-1 function together with FBF to inhibit the activity of the meiotic entry pathways. Given that FBF is present throughout the PZ, spatially restricted SYGL-1 and LST-1 would limit post-transcriptional repression of the three meiotic entry pathways to the distal-most germ cells. Where SYGL-1 and LST-1 are no longer present, FBF would then no longer post-translationally repress meiotic entry, allowing the three meiotic entry pathways to promote meiotic development. Additionally, genetic and molecular data suggest that in the absence of SYGL-1 and LST-1, FBF, through binding to GLD-2 and GLD-3, may actively promote mRNA stabilization and translation, and thereby effectively function in the GLD-2 pathway (Hansen and Schedl 2006; Kimble and Crittenden 2007; Suh et al. 2009; Shin et al. 2017). Thus, an important step in network regulation is FBF partnering with SYGL-1 and LST-1 to repress meiotic entry pathways, and switching to new partners, possibly including GLD-2/GLD-3, to activate meiotic entry. At the tissue level, cells in the distal PZ are stem cells due to the activity of SYGL-1 and/or LST-1 with FBF. When cells become displaced proximally away from the distal tip, the level of SYGL-1 falls below an activity threshold, relieving FBF repression and triggering cells to complete their ongoing mitotic cell cycle and then to undergo meiotic S phase. Further increases in the activity of the three meiotic entry pathways then results in overt transition of cells into early prophase of meiosis I (leptotene).
The switch in GLD-1 pathway regulation, via control of GLD-1 levels, is the best understood of the three meiotic entry pathways (Figure 6). GLD-1 is at a low basal level in the distal-most ∼5–7 cell diameters where LST-1 and SYGL-1 are high and, in conjunction with FBF, this results in gld-1 mRNA destabilization and translational repression (Voronina et al. 2012; Brenner and Schedl 2016; Shin et al. 2017). From ∼7 to 13 cell diameters, GLD-1 levels increase significantly, where LST-1 is absent and as SYGL-1 levels are falling. LST-1 appears to have a greater quantitative contribution to repression of gld-1 mRNA since the lst-1 null causes a greater rise in GLD-1 levels than does the sygl-1 null. Thus, although LST-1 and SYGL-1 are redundant, LST-1 appears to control the strength of repression, while SYGL-1 controls the spatial extent of repression. GLD-1 levels continue to rise significantly, reaching plateau peak levels at ∼25 cell diameters from the distal tip. NOS-3, in parallel with the GLD-2 pathway and possibly in conjunction with FBF, promotes the GLD-1 rise to peak level.
The switch in GLD-2 pathway regulation is less clear. GLD-3 is found throughout the PZ (Eckmann et al. 2002). GLD-2 levels are lower in the distal PZ compared to the proximal PZ, as a consequence of post-translational repression by FBF (Wang et al. 2002; Millonigg et al. 2014). However, unlike GLD-1, this mechanism affects GLD-2 level by only a few fold, and there is no evidence that elevation of distal GLD-2 levels drives meiotic entry, suggesting that GLD-2 regulation may not account for the switch-like change. As described above, FBF may promote GLD-2-GLD-3–mediated translational activation. Thus, an attractive possibility is that the targets of GLD-2-GLD-3 may be subject to FBF-mediated post-transcriptional repression in the presence of SYGL-1 and LST-1, but in the absence of SYGL-1 and LST-1, FBF may act with GLD-2-GLD-3 to promote the post-transcriptional activation of these same targets. For SCFPROM−1, how its activity is regulated is not currently known.
There is additional molecular complexity yet to be resolved in the network at level of FBF. FBF is not equivalent to SYGL-1 and LST-1 (nor to GLP-1 signaling). That is, other genes must function to regulate the switch in larvae since loss of FBF causes a Glp-1–like meiotic entry defect only in late larval and adult stages. For example, other PUF family members (Wickens et al. 2002) and/or FOG-1, a cytoplasmic polyadenylation element binding (CPEB) protein, may function redundantly with FBF during larval development to inhibit meiotic development (Thompson et al. 2005). In addition, FBF is not required to repress GLD-1 accumulation in males since there is no change in distal PZ GLD-1 levels in fbf-1/-2 null double mutant males (Suh et al. 2009). What gene(s) repress GLD-1 accumulation in the distal PZ in males is not known.
Defining Stem Cells
DTC–niche-dependent stem cells are undifferentiated, have proliferation potential, and have high GLP-1 signaling (high LST-1 and/or SYGL-1) and low meiotic entry pathway activity (low GLD-1, GLD-2, SCFPROM−1). These properties are shared with germ cells in glp-1(oz112) tumors (see Appendix). Meiotic entry tumors (e.g., GLD-1 pathway, GLD-2 pathway double mutant) also have undifferentiated germ cells, with proliferation potential, even in the absence of GLP-1 signaling and LST-1 and SYGL-1, and low meiotic entry pathway activity. While these cells could be considered stem cell–like, they may be more similar to cells in wild type that have lost GLP-1 signaling and are completing their ongoing mitotic cell cycle (Figure 2). However, instead of progressing to meiotic S phase as they normally would, they continue mitotic cycling.
In a population-based stem cell system, knowledge of the number of stem cells is important for understanding stem cell system dynamics and for determining how perturbations change the system. Levels of LST-1, SYGL-1, and GLD-1 change dramatically in the PZ and are sufficient for the stem cell fate or meiotic development, respectively. Thus, their relative levels, in principle, define cell fate identity. However, it is not known, for example, what level of SYGL-1 is necessary for the stem cell fate. SYGL-1 accumulation can extend as far as 13 cell diameters from the distal tip, which corresponds to a region containing ∼125–135 cells (Shin et al. 2017). However, this is likely an overestimate for the number of stem cells. After 10 cell diameters, SYGL-1 levels have fallen significantly and, correspondingly, GLD-1 levels have risen significantly (Figure 5 and Figure 6; Brenner and Schedl 2016; Shin et al. 2017; Kocsisova et al. 2019). Additionally, Spike et al. (2018) have shown that RNA-binding proteins LIN-41 and GLD-1 are post-translationally inactivated prior to their degradation; similarly, proximal SYGL-1 may be inactivated prior to degradation. Thus, we propose that the number of stem cells in the young adult hermaphrodite is somewhere between 60 and 80, estimated from population dynamics, and 125–135 estimated from SYGL-1 staining.
Going forward, even though SYGL-1 should not be used categorically and without qualification to define stem cell number, the combination of high SYGL-1 and low GLD-1 levels provides a correlative readout of stem cell identity that can be used in future comparative studies to approximate the number of stem cells over the life cycle, in different genetic backgrounds, and in different environmental conditions.
Control of Progenitor Zone Size and its Relationship to Stem Cell Fate
Genetic manipulations that change the size of the PZ may or may not alter the number of stem cells. The size of the adult PZ, the total number of PZ cells, and the zone length in cell diameters from the distal tip (see above) are the product of multiple processes, including the number of stem cells, rate of mitotic cycling, rate of meiotic entry, and the extent of PZ expansion during larval development, which is regulated by worm physiology (see below). Thus, not all genes that affect PZ size function in the stem cell vs. meiotic development decision. For genes that promote the stem cell fate, mutations that reduce activity are expected to decrease the number of stem cells, and thus should decrease the PZ size. For glp-1(rf-ts) at the pt, as well as other GLP-1 signaling pathway genes (e.g., hop-1), PZ length is shorter and cell number is decreased due to fewer stem cells (Fox and Schedl 2015; Lee et al. 2016; Agarwal et al. 2018). For genes in the meiotic entry pathways, since they act downstream of GLP-1 signaling and downstream of LST-1 and SYGL-1, their loss is not predicted to affect stem cell number, but may affect meiotic entry kinetics and thereby increase the size of the PZ. In gld-2 and gld-3 single null mutants the PZ is longer, has increased cell number and, importantly, the rate of meiotic entry is lower than in wild type, suggesting that conversion from mitotic cell cycling to overt meiotic entry is disrupted (Eckmann et al. 2002; Fox and Schedl 2015). Unexpectedly, in gld-1 and nos-3 null single meiotic entry mutants the PZ is shorter and contains fewer cells, unlike the gld-2 and gld-3 single mutants (Brenner and Schedl 2016). This observation suggests that GLD-1 and NOS-3 have additional functions to promote mitotic cell cycling or PZ expansion during larval development. Mutations or conditions that alter physiology can also affect PZ size. For example, disruption of DAF-2 IGF/insulin-like signaling results in an adult PZ with fewer cells because of a slower cell cycle during PZ accumulation in larval development (see below; Michaelson et al. 2010). However, DAF-2 IGF/insulin-like signaling does not have a direct role in specification of the stem cell fate since reducing the function of daf-2, the sole Insulin Receptor (IR) ortholog, fails to enhance the penetrance of the Glp-1 phenotype of glp-1(rf-ts) pt. Additionally, daf-2(rf) mutant gonads are thinner than wild type, so while the number of PZ cells is decreased, the length of the PZ as measured by distance from the distal tip is similar to wild type (Michaelson et al. 2010). Finally, alterations in germ cell size can conceivably alter the relationship between PZ cell number and distance from the distal tip (Korta et al. 2012). Together, these results illustrate the importance of detailed analysis of the cell cycle, meiotic entry, examination of larval expansion, as well as assessment of the cellular extent of SYGL-1 to provide an understanding of the basis of the altered adult PZ size.
Additional Regulators of the Stem Cell Fate vs. Meiotic Development Decision
Additional genes that promote the stem cell fate are indicated in Table 2 and additional genes that function in entry into meiosis are listed in Table 3. Analysis of these genes is less extensive than those discussed above, so it is uncertain how they function in the regulatory network. Below, we describe the basis for inclusion of genes in one or the other group and discuss several examples.
Table 2. Genes that promote the stem cell fate—enhancer of glp-1 hypomorph.
| Genea,b | Product | Human homolog | Site of action | Reference(s) |
|---|---|---|---|---|
| Cyclin E/CDK2 | ||||
| cye-1 | Cyclin E | CCNE1 | Germ line | Fox et al. (2011) |
| cdk-2 | Cyclin-dependent kinase | CDK2 | Germ line | Fox et al. (2011) |
| RAF-ERK MAP Kinase signaling | ||||
| lin-45 | Mitogen activated protein kinase kinase kinase | BRAF | Germ line | Lee et al. (2007) |
| mek-2 | Mitogen activated protein kinase kinase | MAP2K2 | Germ line | Lee et al. (2007) |
| mpk-1 | Mitogen activated protein kinase | MAPK1 (Erk) | Germ line | Lee et al. (2007) |
| CSR-1 complex | ||||
| ego-1 | RNA-dependent RNA polymerase | — | Germ line | Qiao et al. (1995); Smardon et al. (2000) |
| csr-1 | Argonaut | — | Germ line | She et al. (2009) |
| drh-3 | DEAH/D-box helicase | DHX58 | Germ line | She et al. (2009) |
| ekl-1 | Tudor domain protein | — | Germ line | She et al. (2009) |
| TOR signaling | ||||
| rsks-1 | Ribosomal protein S6 kinase | RPS6KB2 | Germ line | Korta et al. (2012) |
| Miscellaneous | ||||
| hsp-90 | Chaperone | HSP90 | Lissemore et al. (2018) | |
| ego-2 | Protein tyrosine phosphatase, nonreceptor type 23 | PTPN23 | DTC | Qiao et al. (1995); Liu and Maine (2007) |
| atx-2 | RNA-binding protein | ATXN2L | Ciosk et al. (2004); Maine et al. (2004) | |
| iffb-1 | Eukaryotic translation initiation factor | EIF5B | Germ line | Yu et al. (2006) |
Enhancement of glp-1 temperature-sensitive (ts) reduction-of-function (rf) allele, at the permissive temperature (noted in the text as “glp-1(rf-ts) pt”), to a Glp-1–like phenotype in which all germline stem cells enter meiosis; glp-1 ts allele employed depends on study, either bn18, q231, or e2141.
Genes that function in the core Notch signaling pathway that were identified based on enhancement of glp-1 ts are listed in Table 1.
Table 3. Genes that promote meiotic development—enhancer of weak glp-1 gain of function.
| Genea | Product | Human homolog | Evidenceb | Reference(s) |
|---|---|---|---|---|
| Pre-mRNA splicing | ||||
| mog-1 | Yeast PRP16 related splicing factor | DHX38 | 2,4 | Belfiore et al. (2004); Kerins et al. (2010) |
| mog-4 | Yeast PRP2 related splicing factor | DHX16 | 2 | Belfiore et al. (2004); Kerins et al. (2010) |
| mog-5 | Yeast PRP22 related splicing factor | DHX8 | 2 | Belfiore et al. (2004); Kerins et al. (2010) |
| cyn-4 | cyclophilin peptidylprolyl | PPIL2 | 2,4 | Belfiore et al. (2004); Kerins et al. (2010) |
| (aka mog-6) | Isomerases | |||
| prp-17 | Yeast PRP17 related splicing factor | CDC40 | 2,4 | Kerins et al. (2010) |
| prp-19 | Yeast PRP19 related splicing factor | PRPF19 | 1 | Gutnik et al. (2018) |
| teg-1 | CD2 cytoplasmic tail-binding protein | CD2BP2 | 2,4 | Wang et al. (2012) |
| teg-4 | Splicing factor 3b subunit 3 | SF3B3 | 2,4 | Mantina et al. (2009) |
| Ubiquitin-mediated proteolysis | ||||
| pas-5 | 20S proteasome alpha-type five subunit | PSMA5 | 2,4 | Macdonald et al. (2008) |
| sel-10 | SCF E3 ubiquitin ligase | FBXW7 | 1,6 | Pepper et al. (2003a) |
| sel-11 | E3 ubiquitin ligase | SYVN1 | 1,6 | Choi et al. (2010) |
| rfp-1 | E3 ubiquitin ligase | RNF40 | 3 | Gupta et al. (2015) |
| ubr-5 | E3 ubiquitin ligase | UBR5 | 1,6 | Safdar et al. (2016) |
| RNA N6-adenosine methyltransferase | ||||
| mett-10 | Methyltransferase 10 domain containing | METTL16 | 1,2,5 | Dorsett et al. (2009) |
| Miscellaneous | ||||
| puf-8 | Pumilio RNA-binding family member | PUM1/PUM2 | 1 | Racher and Hansen (2012) |
| kin-10 | Casein kinase 2 beta | CSNK2B | 2,4 | Wang et al. (2014) |
| him-17 | THAP domain containing | — | 3 | Bessler et al. (2007) |
For all genes listed, RNAi or double mutant with weak glp-1(gf) at the permissive temperature results in a strong overproliferation phenotype.
For a gene to be included in the table, at least one additional line of experimentation (evidence 1–6, below) must be available that informs on its function in the stem cell vs. meiotic development decision. 1, RNAi or double mutant results partial or complete suppression of a glp-1(rf) at the restrictive temperature; 2, RNAi or double mutant results in a synthetic tumorous phenotype with gld-2 null and/or with gld-3 null; 3, RNAi or double mutant does not result in a synthetic tumorous phenotype with gld-2 null and/or with gld-3 null; 4, RNAi or triple mutant results in a synthetic tumorous phenotype with gld-3 null and glp-1 null; 5, RNAi or triple mutant results in a Glp-1 like phenotype with gld-3 null and glp-1 null; 6, RNAi or mutant suppresses weak lin-12 loss of function.
Additional genes that promote the stem cell fate
A collection of genes that promote the stem cell fate and/or inhibit meiotic development have been identified whose loss of function enhances the Glp-1 phenotype of glp-1(rf-ts) pt (Table 2). Here, enhancement refers to elevation of the penetrance of the Glp-1 phenotype (differentiation of all stem cells). These genes are not essential for the stem cell fate, as single mutants/RNAi has no obvious effect on stem cell fate, suggesting that the identified molecular complexes/pathways participate in processes where there is redundancy.
The mitotic cell cycle regulator CYE-1/CDK-2 was identified in a screen to assess whether cell cycle regulators promote the stem cell fate. Germline-restricted RNAi of cye-1 and cdk-2 caused premature meiotic entry in glp-1(rf-ts) pt, unlike RNAi of other cell cycle regulators (e.g., cdk-1), which caused PZ mitotic cell cycle arrest (Fox et al. 2011; Jeong et al. 2011). CYE-1 acts parallel or downstream of the GLD-1 and GLD-2 pathways since ectopically proliferating cells in GLD-1 GLD-2 pathway double null mutants enter meiosis upon depletion of cye-1 (Fox et al. 2011). CYE-1/CDK-2 has also been reported to function in repression of GLD-1 accumulation in the distal PZ (Jeong et al. 2011). However, this cannot be the only function of CYE-1/CDK-2 as cye-1 RNAi induces meiotic entry even in the absence of GLD-1. A speculative mechanism for how CYE-1/CDK-2 promotes the stem cell fate is through phosphorylation and inactivation of proteins that promote meiotic development, distinct from phosphorylation of proteins that regulate mitotic cell cycle progression. CYE-1/CDK2 has been shown to promote germline stem cell maintenance in the Drosophila ovary, although it is unclear if the mechanism is similar to that described above in C. elegans (Ables and Drummond-Barbosa 2013).
The RAF–ERK pathway promotes the stem cell fate/inhibits meiotic development. Loss of pathway activity results in enhancement of the Glp-1 meiotic entry phenotype in glp-1(rf-ts) pt (Lee et al. 2007; Table 2). Conversely, a germline tumor arises in double mutants of gap-3 (RAS GAP) and puf-8 (PUF RNA-binding protein) that is dependent on let-60 RAS and mpk-1 ERK and is accompanied by inappropriate activation of MPK-1 ERK throughout the germ line (Vaid et al. 2013). These RAF–ERK functions appear to be specific to late larvae/adults, barring earlier maternal rescue. Attempts to place RAF–ERK signaling in the stem cell fate vs. meiotic development decision has yielded paradoxical results. The gap-3; puf-8 tumor requires GLP-1 signaling, suggesting that it acts upstream the GLP-1 pathway (Vaid et al. 2013); however, gap-3 puf-8 tumors do not show ectopic SYGL-1 accumulation, indicating that the tumors do not have inappropriate GLP-1 signaling output (A. Mohammad, personal communication). How RAF–ERK inhibits meiotic development is unknown.
The CSR-1 complex, which acts at least in part through CSR-1 Argonaute-associated 22G small RNAs, promotes the stem cell fate (Smardon et al. 2000; She et al. 2009). This function appears to act in parallel with or downstream of the GLD-1 and GLD-2 pathways (Vought et al. 2005). The CSR-1 complex has multiple functions in germline development, where the molecular mechanisms are just beginning to be understood. These include transcriptional repression of unpaired chromosomes and chromosomal regions through histone H3 lysine nine demethylation (She et al. 2009), RNAi activity (Yigit et al. 2006), promoting the correct pattern of protein-coding gene expression (Claycomb et al. 2009; Almeida et al. 2019), histone mRNA processing (Avgousti et al. 2012), and ensuring normal morphology of P granules, which are perinuclear hubs for regulation of mRNA translation and stability (Vought et al. 2005; Updike and Strome 2009). One possibility is that the CSR-1 complex plays an active role in the stem cell fate, inhibiting the expression of a negative regulator or promoting expression of a positive regulator, through one or more of the above mechanisms. Alternatively, since the CSR-1 complex also protects germline-expressed genes from silencing via Piwi-interacting RNA–mediated genome surveillance (Almeida et al. 2019), the enhancement of glp-1(rf-ts) pt may be a consequence of unopposed Piwi-interacting RNA–mediated silencing of important stem cell fate genes in the CSR-1 complex mutants.
The sole C. elegans p70 S6 kinase ortholog RSKS-1 acts germline-autonomously to promote the stem cell fate as indicated by enhancement of the Glp-1 meiotic entry phenotype in combination with rsks-1(lf) (Korta et al. 2012; Roy et al. 2018). Loss of rsks-1 also suppresses the penetrance of tumor formation caused by glp-1(gf). The enhancement of glp-1(rf-ts) pt phenotype formed the basis of a large-scale RNAi screen that identified several genes that act in a genetically linear pathway with rsks-1 to enhance glp-1(rf-ts) pt (Roy et al. 2018). The exact role(s) of rsks-1 and the other enhancers in the stem cell vs. meiotic development decision remain to be determined.
Additional genes that promote meiotic development
A collection of genes that promote meiotic development and/or inhibit the stem cell fate have been identified whose loss of function enhances the tumorous phenotype of weak glp-1(gf-ts) mutants (Table 3; also see Appendix). Single mutants/RNAi, in almost all cases, confer no obvious meiotic entry defective phenotype, suggesting that they likely participate in redundant processes.
Genes throughout the pre-mRNA splicing cascade function in the stem cell fate vs. meiotic development decision; >35 splicing factor genes, when mutant or knocked down by RNAi, are enhancers of weak glp-1(gf-ts) pt and are synthetic tumorous with gld-3 null (Table 3; Puoti and Kimble 1999, 2000; Belfiore et al. 2004; Mantina et al. 2009; Kerins et al. 2010; Zanetti et al. 2011; Wang et al. 2012). The splicing factors may act in the GLD-1 pathway, downstream (or in parallel) to GLP-1 signaling, as the synthetic tumorous phenotype with gld-3 null mutants is epistatic to glp-1 null. Splicing factor mutants are also synthetic tumorous with gld-2 null; however, unlike with gld-3, this overproliferation is dependent on GLP-1 signaling (Kerins et al. 2010; Wang et al. 2012). The reason for the difference in GLP-1 dependence between the splicing factor genes with gld-2 vs. gld-3 is unclear, but may be a consequence of redundancy of GLD-2 and GLD-4/GLS-1 polyA polymerase activity (Millonigg et al. 2014). The mechanism by which the pre-mRNA splicing cascade promotes meiotic development or inhibits the stem cell fate is unknown. One model is that decreased splicing, missplicing, or defective alternative splicing is not tolerated by certain genes that function in the decision. Another model is that reduced splicing factor activity indirectly disrupts a distinct regulatory process, such as cytoplasmic mRNA stability or translational regulation, for genes that function in the decision. For example, some splicing factors associate with the pre-mRNA in the nucleus and remain associated postsplicing in the cytoplasm, such as the exon junction complex (Le Hir and Seraphin 2008). Reduced activity may alter the level of such bound factors on mRNAs, which would result in abnormal expression patterns and thereby misregulate the decision.
Components of the ubiquitin-mediated proteolysis system function to inhibit the stem cell fate, based on enhancement of the overproliferation phenotype of weak glp-1(gf-ts) pt. Four are different classes of E3 ubiquitin ligases (Table 3), which act as substrate specificity subunits for ubiquitination. The different classes suggest that there are distinct protein substrates and/or different recognition motifs within a given substrate. The Notch ICD is a well-known target for ubiquitin-mediated degradation, which acts at least in part through the C-terminal PEST destabilization region (Weng et al. 2004). GLP-1(ICD) has a PEST region similarly positioned as in other Notch homologs. SYGL-1 and LST-1 are additional candidate substrates for ubiquitin-mediated degradation. For these proteins, loss of a protein destabilization mechanism may result in only a modest increase in stem cell number/PZ size in the wild type, given the spatial restriction of ligand-dependent generation of ICD and restriction of sygl-1 and lst-1 transcription. The germline substrates for the four E3 ligases are not known, but since sel-10, sel-11, and ubr-5 also genetically interact with lin-12, these may act on one or more core Notch signaling components (Greenwald and Kovall 2013). Indeed, SEL-10 FBW7 interacts with LIN-12 and SEL-12 (Hubbard et al. 1997; Wu et al. 1998), and SEL-10 promotes degradation of LIN-12(ICD) in some cells of the vulval precursor cell lineage (Deng and Greenwald 2016).
Splicing factors have also been reported to promote the degradation of GLP-1(ICD) (Gutnik et al. 2018). To visualize nuclear GLP-1(ICD), as well as plasma membrane GLP-1 that has not undergone ligand-dependent cleavage (see above), GFP was inserted just N-terminal to the PEST region in the GLP-1 cytoplasmic domain. In L4 stage worms, nuclear GFP fluorescence was restricted to the distal-most ∼5 cell diameters, consistent with the spatially restricted nascent sygl-1 transcripts observed by single-molecule fluorescent in situ hybridization in adults (Lee et al. 2016). In adults, nuclear GFP signal was below the limit of detection. Following RNAi of splicing factor genes (e.g., prp-19, mog-4), as well as the proteasome components pbs-1 and E1 conjugating enzyme uba-1, nuclear GFP signal was observed in the adult, suggesting stabilization of GLP-1(ICD). Under conditions where nuclear GFP fluorescence was observed, the nuclear GFP signal was often similar to or brighter than the surrounding plasma membrane signal; this is unexpected given the idea that ligand-dependent cleavage to generate GLP-1(ICD) is rate limiting, in the context of excess intact GLP-1 at the plasma membrane. However, downregulation of GLP-1(ICD) cannot be the only function of the splicing factors in the stem cell fate vs. meiotic entry decision since loss of splicing factor activity in the gld-3 null background results in a tumorous phenotype even in the absence of GLP-1 signaling.
Germline Stem Cell System During Larval Development
Larval progenitor zone accumulation in replete, continuous development conditions
Over the course of larval development, the number of cells in the PZ increases to generate the adult PZ described above that supports adult fertility and fecundity. Here, we use the term “accumulation” to describe this increase in cell number. Two factors contribute to PZ accumulation: cell fate, which dictates mitotic competence, and cell cycle rate. As described above, due to the way the C. elegans germ line develops, these two factors are genetically separable but are linked when considering PZ accumulation.
The number of cells in the PZ increases during larval stages, and the pace and timing of this accumulation has several important ramifications for the adult. Figure 7A shows PZ accumulation over time (Hirsh et al. 1976; Pepper et al. 2003b; Killian and Hubbard 2005), from the two cells of the newly hatched L1 larva to ∼200–250 PZ cells in the adult over the course of ∼3 days, subject to exact temperature and nutrient conditions. The accumulation dynamic of the PZ in larval stages determines the size of the PZ when worms reach reproductive maturity in the adult, affects the number of gametes produced, and thus affects fecundity. PZ accumulation is also critical to establish the correct distal-proximal pattern during germline development (see Appendix). Prior to “initial meiosis” (Pepper et al. 2003b), the time at which proximal-most germ cells first enter the meiotic pathway in the L3 (Figure 7), all germ cells can be considered as stem/progenitor cells. Initial meiosis is the event that establishes the border between cells in the PZ (distal) and leptotene cells (proximal), and thus establishes the polarized germline assembly line–like features of meiotic prophase progression. The PZ-leptotene border is initially sharp in larvae, occurring over the distance of one cell diameter (Hansen et al. 2004a), contrasting with the extended meiotic entry region in the adult (see above). Here, we compare the larval and adult PZ, describe larval stem cell identity and PZ accumulation, and discuss genetic and environmental factors that affect larval PZ accumulation.
Figure 7.
Dynamic changes in the germline stem cell system during development and adulthood and in altered environments. Approximate number of progenitor zone (PZ) nuclei per gonad arm over time. (A) PZ accumulation in standard laboratory conditions. Solid line: PZ accumulation in larval stages, homeostasis in early adult, and decline in aging. Dashed line: PZ accumulation after Sh1 (distal sheath cell pair) ablation. (B) Comparison of PZ accumulation in standard laboratory conditions (solid line) vs. poor conditions or reduced nutrient signaling (dashed lines). In A and B, time of initial meiosis and estimated trajectory for total PZ cells produced (including cells that enter meiotic prophase starting in the mid-L3; Berry et al. 1997) is represented by the dotted arrow. (C) Comparison of PZ accumulation in continuous development vs. developmental arrest (L1 arrest/diapause, dauer, and ARD; see text). L1, L2, etc. indicates first, second, etc. larval stages; D1, D2, etc. indicates adult age progression, e.g., “day 1” adult. Color code: blue, accumulation phase; gray, homeostasis; orange, decline; tan, continued quiescence; green, regrowth after ARD arrest and recovery. ARD, adult reproductive diapause; GC, germ cell.
Comparison of larval and adult stem cell system
The cellular and molecular regulation of the stem cell fate vs. meiotic development decision during larval stages is likely similar to the adult, but it has not been investigated in as much detail. For example, although it is well established that DTC–GLP-1 pathway signaling is required at all stages, a complete time course of the relative proportion of cells in the PZ that are actively responding to GLP-1 via LST-1 and SYGL-1 has not yet been reported. It will be of interest to determine how each pool within the PZ (Figure 2) is established during the dynamic larval PZ growth period, how the relative proportions of cells in each pool affect total PZ accumulation, and how the different perturbations that limit larval PZ accumulation affect these proportions.
Several features of the accumulating larval PZ differ from the homeostatic condition in the early adult. The average MI in the PZ is higher in larval stages (Michaelson et al. 2010; Korta et al. 2012; Narbonne et al. 2015; Roy et al. 2016), consistent with a faster cell cycle. The higher average MI in larval stages does not depend on whether cells are destined to become sperm vs. oocytes. In addition, larval cell cycles are associated with a slightly lower average DNA content, suggesting that the rates of S and G2 progression may not be equivalent in larvae and adults. The accumulation rate is not strictly limited by developmental stage since adult germ cells can accumulate at larval rates under conditions where the adult PZ is reestablished after depletion and arrest (see below, section ARD) (Roy et al. 2016). What accounts for larval vs. adult cell cycle differences remains unknown. One possibility is that cell crowding may contribute (Atwell et al. 2015). Physiological status affects larval and adult PZ cells differently (see below). PZ cells also differentially require members of several partially redundant gene pairs (e.g., fbfs, Crittenden et al. 2002; skr-1/-2, Mohammad et al. 2018; hop-1/sel-12, Agarwal et al. 2018), suggesting the possibility that large-scale genetically regulated changes occur in the PZ as animals transition through larval stages into adulthood. Finally, the metabolic landscape of larvae differs from reproductive adults that are manufacturing yolk-rich oocytes. In particular, the overall metabolic rate of larvae is higher and lipid metabolism of adults is altered to facilitate oocyte production (yolk synthesis) and fertilization (prostaglandin synthesis) [see reviews Braeckman et al. (2009) and Watts and Ristow (2017)].
The niche and gonad architecture also differ between larval and adult stages. The larval hermaphrodite DTC actively migrates. Although alterations in the DTC migration route do not interfere with fertility (Hedgecock et al. 1987, 1990; Antebi et al. 1998), sufficient early migration is necessary for timely initial meiosis and patterning (see Appendix, also see below). The larval DTC morphology is relatively simple and it enwraps a relatively small group of distal-most cells until the late L4, when the plexus that characterizes the adult DTC is elaborated (Byrd et al. 2014). The larval rachis is initially much narrower than the adult, and during the L4, the distal gonad changes shape as it fills with germ cells: the rachis expands, the distal gonad grows circumferentially, and the entire gonad stretches in the anterior-posterior direction (Atwell et al. 2015). Finally, the folds that can form in the adult hermaphrodite gonad are not present in larvae (Seidel et al. 2018).
Primordial germ cells and L1 germ cells
Several recent reviews cover the early separation of germline and somatic fates in the embryo and the maintenance of germ cell fate (Lehmann 2012; Wang and Seydoux 2013; Strome and Updike 2015). Here, we focus on germline stem/progenitor status. Similar to other animals, C. elegans PGCs are largely transcriptionally silent during embryogenesis (Schaner et al. 2003), and embryonic PGCs arrest in G2 (Fukuyama et al. 2006). In certain mutants, PGCs undergo inappropriate cell divisions during embryogenesis. This occurs upon loss of nanos-related genes nos-1 and nos-2 or certain PUF family members (fbf-1, fbf-2, puf-6, puf-7, and puf-8) (Subramaniam and Seydoux 1999; Lee et al. 2017). However, these are not prematurely activated germline stem cells. Rather, an autonomous defect in germ cell fate specification in nos mutant PGCs leads to a retention of oocyte transcripts and inappropriate expression of factors that drive somatic fate and cell cycle (Lee et al. 2017).
PGCs must first transition out of embryonic quiescence before establishing the stem cell pool. At hatching, the gonad primordium comprises the two PGCs (Z2 and Z3) flanked by two SGPs (Z1 and Z4), all surrounded by a basement membrane (Sulston and Horvitz 1977). If food and the SGPs are present (Hirsh et al. 1976; Kimble and Hirsh 1979; Kimble and White 1981), Z2 and Z3 exit G2 arrest and begin dividing midway through the L1 stage. The SGPs make gap junctions with the PGCs and these are required for germ cell proliferation (Starich et al. 2014; see below). The food signal is mediated by the insulin signaling pathway (see below, section L1), but the mechanism underlying the requirement for SGP contact for PGC proliferation in the L1 is unknown. Additional events accompany the transition from quiescence to proliferation in the L1. Within 1–2 hr of hatching on food, RNA Pol II activity becomes elevated in Z2 and Z3 (as indicated by staining with the H5 antibody that recognizes phospho-Ser2 of the C-terminal domain of Pol II), and division of Z2 and Z3 ensues at ∼4–6 hr posthatching on food (Hirsh et al. 1976; Kimble and Hirsh 1979; Butuči et al. 2015). After RNA Pol II activity is detected, evidence of DNA damage is observed (phospho-CHK-1 and RAD-51 foci), which is then repaired prior to the first PGC division (Butuči et al. 2015). Transient DNA breaks are thought to quickly decompact the genome and allow for large-scale transcriptional activation (Wong et al. 2018). PGC chromatin compaction, perhaps a mechanism to safeguard the genome during long quiescence, is first established embryonically and later relaxed (after L1 feeding), and both of these processes apparently involve the TOP-2 topoisomerase II. While DNA damage seems risky in cells that will give rise to the next generation, the risk may be mitigated by the opportunity for homologous repair from sister chromatids in these G2-arrested germ cells.
The “stem cell” status of germ cells during the first one to two cell cycles is unclear. In glp-1 zygotic null mutant animals, Z2 and Z3 undergo one to two divisions and then prematurely differentiate to generate an average of four to eight germ cells (Austin and Kimble 1987). Several aspects of this observation remain unresolved. What prevents direct differentiation of Z2 and Z3 in the absence of glp-1? One possibility is the presence of maternal GLP-1. This possibility is difficult to test due to the requirement for glp-1 in the maternal germ line (Austin and Kimble 1987) and in a series of early embryonic cell fate decisions (Priess 2005). However, strong temperature-sensitive mutants shifted in embryogenesis after the maternal glp-1 requirement in the early embryo produce the same germline glp-1 phenotype as the null, arguing against the maternal contribution hypothesis (Austin and Kimble 1987). Another possibility is that the PGCs must undergo one to two divisions before acquiring competence to differentiate. This would be consistent with the terminal division model postulated for the adult germline stem cell system described above. Finally, it is possible that Z2 and Z3 are fundamentally different from later GLP-1–responsive stem cells, and that they must divide to become competent to respond to GLP-1.
Early larval PZ accumulation also requires certain innexins that form gap junction–like connections between the somatic gonad and germ line. Redundant inx genes inx-8 and inx-9 in the soma and inx-14 and inx-21 in the germ line are critical for germ cell proliferation (Starich et al. 2014). A combination of mosaic analysis and heterologous gene expression indicated that, while DTC expression of inx-8 is sufficient to rescue the proliferation defect, it is not necessary since sheath-expressed inx-8 can also rescue. Interestingly, the ∼8 germ cells that form in inx-8 inx-9 double mutants are not competent to differentiate even in the absence of glp-1. Further, the proliferation defect is epistatic to germline stem cell tumors and to meiotic entry tumors alike. Thus, these junctions are critical for both proliferation and differentiation. The identity of the relevant molecules that traverse these junctions is unknown.
Rapid accumulation of germline progenitors in L2–L4 establishes germline pattern
Anatomical, genetic, and physiological factors together affect PZ accumulation during larval stages. After the first rounds of division, early larval germ cells are considered stem cells since they require GLP-1 Notch signaling to remain undifferentiated. In the late L1 and early L2, the DSL ligand LAG-2 is required in four cells to prevent premature differentiation: the two DTCs (Z1.aa and Z4.pp) and two central somatic gonad cells (Z1.pp and Z4.aa) (Wilkinson et al. 1994; Pepper et al. 2003b). While DTC ablation or loss of glp-1 each causes all germ cells to enter meiosis, the early requirement for LAG-2 in non-DTC cells (Pepper et al. 2003b) largely accounts for the temporal difference in meiotic entry between early DTC ablation (normal timing of meiotic entry in the L3) and the glp-1 null mutant (early meiotic entry in the L2) (Kimble and White 1981; Austin and Kimble 1987). Gap junctions that remain between the DTC and germ cells after DTC ablation may also contribute to difference in timing of meiotic entry after DTC ablation relative to the glp-1 null (Starich et al. 2014). At the end of the L2, the somatic gonad undergoes a morphological rearrangement: one DTC remains at each end of the two (anterior and posterior) gonad arms, and the remaining somatic gonadal cells coalesce in the center to form the somatic gonad primordium (Kimble and Hirsh 1979). The germ cells continue to divide, generating ∼20 germ cells per gonad arm by the end of the L2 (Hirsh et al. 1976). During the L3 and L4 stages, the progenitor pool accumulates rapidly, giving rise to additional progenitors as well as cells that enter the meiotic pathway starting from the mid-L3 stage (Figure 7) (Pepper et al. 2003b; Hansen et al. 2004a; Killian and Hubbard 2005). Thus, the morphological rearrangement of the somatic gonad localizes the ligand source for stem cells exclusively to the distal tip of each gonad arm and establishes the subsequent pattern of germline development.
Concurrent with the rapid expansion of the PZ in the L2 and L3, each arm of the hermaphrodite gonad is led by its DTC toward the anterior and posterior of the animal. Importantly, the distance reached by the DTC is the combined result of the intrinsic DTC migration plus the force of additional biomass from proliferating germ cells within the gonad (imagine air filling a long balloon). Anatomical and genetic manipulations (Killian and Hubbard 2004, 2005; Voutev et al. 2006; Voutev and Hubbard 2008; McGovern et al. 2009) together with computer modeling studies (Atwell et al. 2015; Hall et al. 2015) underscore the importance of these combined spatial and temporal aspects of gonad development for germline patterning (see Appendix). Since alterations in larval somatic or germline morphogenesis can lead to surprising outcomes in the adult, interpretation of adult phenotypes requires consideration of events in all preceding stages.
The distal pair of somatic gonadal sheath cells (Sh1) is required for optimal larval PZ accumulation (Figure 7A; McCarter et al. 1997; Killian and Hubbard 2004). While Sh1 cells contact the PZ from the time of their birth in the early L3, Sh1 nuclei are centered over the pachytene region by the early adult stage. The thin Sh1 cell body covers much of the dorsal arm of the gonad, closely following the contours of the germ cell membranes, and sends processes distally into the proximal part of the PZ (Hall et al. 1999). Ablation of Sh1 alone, but not the other pairs of sheath cells (Sh2–5), impairs PZ accumulation (Killian and Hubbard 2004), but only slightly delays meiotic entry, so no proximal latent niche–dependent germline stem cell tumor forms (see Appendix). If, however, Sh1 is ablated in the glp-1(ar202 gf) mutant background at the pt, where meiotic entry is independently slightly delayed due to elevated Notch signaling but would not otherwise cause tumor formation, a latent niche tumor invariably forms (Killian and Hubbard 2005). Thus, manipulations that slow larval germ cell cycle progression but do not otherwise grossly interfere with somatic development, cell cycle competence, or stem cell fate (such as defects in insulin signaling, see below) can, counterintuitively, enhance the penetrance of tumor formation of the proximal latent niche–dependent type (see Appendix). The molecular basis for the Sh1-germline interaction that underlies its role in larval PZ accumulation is not known, although ribosomal RNA processing is important, as revealed by the requirement for pro-1 in the sheath lineage for robust PZ accumulation. PRO-1 is a WD repeat containing protein that, like its yeast ortholog IPI3, is important for removal of the internal transcribed spacer two from preribosomal RNA. Other aspects of ribosome biogenesis are likely also important since loss of PRO-2 NOC2L and PRO-3 SDA1 affect the sheath lineage and/or cause a Pro phenotype (Killian and Hubbard 2004; Voutev et al. 2006).
Environmental Modulation of the Germline Stem Cell System
Environmental modulation of larval PZ accumulation
Several conserved signaling pathways modulate larval PZ accumulation in response to the animal’s environment and/or physiological state. Since reproduction is costly, correlating physiological state with the PZ is likely an important mechanism to match reproductive output with available resources. Accordingly, poor conditions impede PZ accumulation (Figure 7B). This strategy makes evolutionary “sense,” considering the patchy distribution of C. elegans food sources in the wild (Schulenburg and Félix 2017). Although the situation is far more complicated in the wild, even in the laboratory where worms are typically grown on monospecific lawns of Escherichia coli bacteria, the environment provides information, including status and changes in temperature, osmolarity, quality and quantity of bacteria, and the density of nearby worms vying for resources. Because the larval PZ is sensitive to subtle changes in the animal’s environment, and because the regulation of cell cycle and cell fate can be experimentally separated, the larval PZ is an attractive model for investigating how whole-animal physiology affects stem cells. As in other animals, C. elegans nutritional status is detected both neuronally (via external cues) and metabolically (via consumed diet). Together, these alter a network of behavioral, signaling, and cell-intrinsic responses in the animal (Bargmann 2006; Fielenbach and Antebi 2008; Braeckman et al. 2009; Avery and You 2012; O’Donnell et al. 2018; Rhoades et al. 2019). In addition, body cavity neurons monitor the internal physiological state (Witham et al. 2016). Here we use the term “physiological state” to encompass changes that occur in the animal in response to the sensory or dietary milieu, or both. While much remains to be discovered, we summarize recent progress on the effects of major conserved signaling mechanisms that link physiological state to larval PZ accumulation, and the interaction of these pathways with food quality and quantity.
Insulin:
The core C. elegans insulin signaling pathway that regulates dauer and many other biological processes (Murphy 2013) promotes robust larval PZ accumulation. As in the dauer decision, DAF-2 IR activity interferes with the transcription factor DAF-16 FOXO to promote PZ accumulation (Figure 8). The current model is that insulin signaling does not affect the GLP-1 Notch-mediated cell fate decision, but rather promotes germ cell G2 progression (Michaelson et al. 2010; Roy et al. 2016), and while larval germ cell cycle is most sensitive to daf-2 activity, it also can promote cell cycle progression in the adult (Narbonne et al. 2015). For larval PZ accumulation, the germ line is the primary, although not sole, focus of activity for DAF-16 FOXO downstream of DAF-2 IR. Notably, daf-16 is not required in neurons nor in the intestine, indicating that the effect on the larval germ line is anatomically distinct from the roles of daf-16 in the dauer decision and in life span (Michaelson et al. 2010).
Figure 8.
Physiological pathway control of stem cell fate and/or cell cycle in larvae. Signaling pathways: DAF-2 insulin-like signaling pathway (top), TOR (middle), and DAF-7 TGFβ (bottom) promote mitotic cell cycle progression (blue) and/or stem cell fate (green). Gene products that inhibit these activities shown in red.
Two of the 40 C. elegans genes that encode insulin-like ligands, ins-3 and ins-33, promote larval PZ accumulation, but additional agonist ligands are likely involved since daf-2(e1370), a canonical rf allele used in daf-2 studies, causes a more severe phenotype than does loss of both ins-3 and ins-33 (Michaelson et al. 2010). Consistent with the model that different INS ligands mediate different tissue-specific and life history effects of DAF-2 (Murphy 2013; Zheng et al. 2018), neither ins-3 nor ins-33 influences daf-2 targets in the intestine nor daf-2–mediated timing of reproductive output in aged animals (Michaelson et al. 2010). Given the similar effect of Sh1 ablation and daf-2(e1370) on the larval PZ, an attractive hypothesis was that Sh1 produces insulin ligands to activate DAF-2 in the germ line; however, depletion of daf-16 does not suppress the PZ accumulation defect caused by Sh1 ablation, although it suppresses the PZ accumulation defect caused by reduced insulin signaling. Moreover, neither ins-3 nor ins-33 appear to be expressed in Sh1. Therefore, it is unlikely that the gonadal sheath alone mediates the effects of insulin signaling on the larval germ line (Michaelson et al. 2010). Several autophagy genes represent an additional input to DAF-2–regulated larval germline cell cycle control. In particular, BEC-1, the C. elegans Beclin1 ortholog, acts germline-nonautonomously to promote cell cycle progression via DAF-2 IR, in this case dependent on DAF-18 PTEN and the transcription factor SKN-1 Nrf (Ames et al. 2017). The relevant targets of DAF-16 and SKN-1 that regulate cell cycle in the larval germ line remain to be determined, as do the relevant environmental or developmental factors that regulate ins genes and their products.
TOR:
The activity of TORC1, a conserved protein complex, and its downstream targets are implicated in cellular responses to amino acids and growth factors (Saxton and Sabatini 2017). C. elegans orthologs are implicated in larval PZ accumulation, including let-363 TOR, daf-15 RAPTOR, and their downstream positive effectors ife-1 eIF4E and rsks-1 p70 S6K (Blackwell et al. 2019; Lapierre and Hansen 2012) (Figure 8). The pathway appears to act germline-autonomously to promote larval germline cell cycle (Korta et al. 2012; Roy et al. 2016), and this function is likely parallel to DAF-2 IR since it is independent of DAF-16 FOXO. Furthermore, the role of RSKS-1 in larval PZ accumulation is distinct from its role in life-span regulation since it shows no dependence on genes that mediate the rsks-1 mutant life-span extension phenotype such as pha-4 FOXA, egl-9 proline hydroxylase, or aak-2 AMPK (Korta et al. 2012).
Genetic analysis of the TORC1 pathway in C. elegans is complicated by pleiotropy and redundancy. Pleiotropy includes larval arrest and slow-growth phenotypes, although these can be bypassed by restricting RNAi or loss of gene activity to the germ line. Redundancy is also present as five genes encode eIF4Es (Keiper et al. 2000), although ife-1 has the most marked effect on the larval PZ (Korta et al. 2012). Reducing let-363 TOR activity also affects the TOR/RICTOR complex, TORC2. Some components of the TORC1 signaling pathway found in other organisms are not yet identified by sequence or functional orthologs in C. elegans, such as the GTPase-activating proteins TSC1 and TSC2, although these functions may be performed by other GTPase-activating proteins in C. elegans (Blackwell et al. 2019). Nevertheless, similar to other systems, TORC1 appears to promote the activity of both eIF4E and S6K since loss of ife-1 and rsks-1 cause an additive PZ accumulation defect that together is similar to the germline phenotype of let-363 TOR and daf-15 RAPTOR (Korta et al. 2012).
In addition to its role in promoting the larval cell cycle, RSKS-1 S6K also promotes the stem cell fate and/or opposes meiotic entry, a role that is not shared by other components of the TORC1 pathway. Loss of rsks-1 markedly enhances the penetrance of the Glp-1 phenotype of glp-1 (rf-ts) pt (Table 2). Like the effect of rsks-1 on PZ accumulation, the cell fate role for rsks-1 is germline autonomous and depends on threonine 404, a highly conserved residue that is a site of TOR phosphorylation in other systems (Korta et al. 2012; Roy et al. 2018). However, neither reduced let-363 TOR nor ife-1 eIF4E enhance the Glp-1 phenotype, even though PZ accumulation in these single RNAi/mutant cases is impaired similarly to loss of rsks-1. One possible explanation for why reduced let-363 or ife-1 loss does not enhance and glp-1 (rf-ts) pt like rsks-1 loss is that the effects of TORC1 and/or eIF4E on GLP-1 Notch-mediated cell fate decision are masked by pleiotropy. Another possibility is that rsks-1 S6K has a separate threonine 404–dependent function that is required to regulate GLP-1 Notch signaling. The relationship between the roles of rsks-1 S6K in germ cell fate and proliferation is not known.
TGFβ:
The DAF-7 TGFβ pathway (known as the “dauer TGFβ pathway” as opposed to the Sma/Mab pathway; Gumienny and Savage-Dunn 2013) promotes larval PZ accumulation. Analysis of this pathway revealed the existence of a neuron-to-niche signaling mechanism that relays information about the environment to larval germline stem cells (Dalfó et al. 2012; Pekar et al. 2017) (Figure 8). Unlike insulin and TOR signaling, the TGFβ pathway acts germline-nonautonomously and via GLP-1 Notch to promote PZ accumulation (although it may have an additional glp-1–independent role in meiotic entry–defective tumors) (Dalfó et al. 2012; Pekar et al. 2017). The expression of daf-7 TGFβ in ASI neurons is elevated by feeding after starvation (Ren et al. 1996; Schackwitz et al. 1996), and these neurons are required for the effect of daf-7 TGFβ on the germ line. Certain nutritionally relevant roles for daf-7 TGFβ, such as feeding behavior, satiety, and metabolism, require daf-1 TGFβ receptor (TGFβR) activity in RIM and RIC interneurons. In contrast, neuronal daf-1 TGFβR does not influence the larval PZ. Instead, results from anatomical, genetic, and environmental manipulation experiments (Dalfó et al. 2012; Pekar et al. 2017) are consistent with a model in which favorable conditions (abundant food and low population density) cause ASI-produced DAF-7 TGFβ to act in a neuroendocrine manner to activate DAF-1 TGFβR on the DTC (Dalfó et al. 2012). There, daf-1 TGFβR activity inhibits a transcriptional repressor complex that includes the DAF-3 repressor SMAD. DAF-3 can bind a 25 bp region of the lag-2 promoter that is required for the response to daf-7 TGFβ (Pekar et al. 2017). Thus, when environmental conditions are poor, lag-2 is expressed in the DTC at lower levels than when conditions are good. In this way, the size of the stem cell pool is modulated by the environment through a two-step relay: from ASI neuron-produced TGFβ to TGFβR signaling in the DTC, and then from DTC-produced LAG-2 to GLP-1 Notch signaling in the germ line (Figure 8).
A general concept that emerges from these studies is that cell fate specification mechanisms, such as those governed by Notch, are subject to modulation by the animal’s physiology. During early development, cell fate specification decisions must be made quickly and irreversibly to ensure the proper order of subsequent developmental events that rely on their timely and accurate resolution. However, during continuous inductive signaling events, such as the niche-to-stem cell signaling that promotes germline stem cell fate over an extended time period, this same pathway can be modulated by the organism’s physiological state.
Food quantity and quality:
Sensory activity, feeding, and metabolism influence multiple aspects of the worm’s life history, including growth, reproduction, and aging. Life history traits are also subject to differences in qualitative aspects of the ingested bacterial diet, the mode of delivery of bacteria (e.g., solid/liquid; Çelen et al. 2018), bacterial metabolites, and the worm’s external and internal microbiota. The reader is directed to several reviews for additional information (Braeckman et al. 2009; Schulenburg and Félix 2017; Watts and Ristow 2017; Zhang et al. 2017; Ezcurra 2018).
The germ line is particularly sensitive to diet. Two examples highlight this sensitivity in the context of two common laboratory E. coli diets: OP50 and HT115. OP50, the common laboratory diet, is a uracil auxotroph derived from E. coli B (Brenner 1974), while HT115 is a strain optimized for RNAi feeding experiments (Timmons et al. 2001) and is derived from E. coli K12. The germline phenotype of nhr-114 mutants raised on these two bacteria exhibit striking differences that were linked to differences in E. coli tryptophan metabolism (Gracida and Eckmann 2013). Mutations in genes encoding cytidine deaminases also confer bacterial diet–specific germline phenotypes. In this case, they are linked to E. coli uridine metabolism (Chi et al. 2016). In both cases, worms bearing the relevant mutations were sterile on OP50 and fertile on HT115.
Diet also affects wild-type C. elegans larval PZ accumulation. A reduced (but not arrest-inducing; see below) concentration of E. coli OP50 slows germ cell cycle progression and larval PZ accumulation (Korta et al. 2012) (Figure 7B). Similarly, cell cycle progression is slower in mutants that restrict food consumption, such as eat-2, which interferes with efficient feeding by disabling the nicotinic acetylcholine receptor; or in mutants that limit protein uptake, such as pept-1, which interferes with oligopeptide transport (Korta et al. 2012). When combined with glp-1(rf-ts) pt, these perturbations further impair PZ accumulation, but they do not cause all PZ cells to enter the meiotic pathway, suggesting that they act primarily on PZ cell cycle rather than cell fate (Korta et al. 2012). However, as noted above, food abundance can also affect the stem cell fate decision via the neuron-to-niche TGFβ regulation of lag-2 expression (Pekar et al. 2017).
Genetic manipulations can uncouple PZ accumulation from bacterial abundance, underscoring the importance of signaling pathways in the response to dietary resources and challenging the model that diet simply provides chemical building blocks necessary for proliferation of germ cells. In certain signaling mutants, food abundance is no longer correlated with PZ accumulation. For example, despite low food abundance and presumably fewer chemical building blocks, the PZ nevertheless accumulates normally in mutants lacking the DAF-3 repressor SMAD (Dalfó et al. 2012). Similarly, although rsks-1 mutants accumulate fewer PZ cells than the wild type, in low food conditions rsks-1 mutants still accumulate a greater number of PZ cells than the wild type (Korta et al. 2012). In genetic situations that allow germ cells to proliferate despite reduced food abundance, sufficient metabolites are likely available for the production of new cells or the necessary metabolites may be scavenged from other cells. Mechanisms that govern the allocation of nutrients or chemical building blocks to specific tissues remain to be discovered.
Although a systematic analysis is lacking, several specific dietary components are important for PZ accumulation. As mentioned above, intestinal oligopeptide transport is important for PZ accumulation and for fecundity (Korta et al. 2012; Spanier et al. 2018), implicating dietary protein. Bacteria also provide certain folates and folate precursors that promote germ cell proliferation and fertility. These functions require the folate receptor ortholog FOLR-1 in C. elegans, possibly independent of the role of folate as a vitamin (which requires the reduced folate carrier FOLT-1) and independent of one-carbon metabolism (Austin et al. 2010; Chaudhari et al. 2016).
Steroid signaling:
Nuclear hormone receptors govern many aspects of C. elegans biology, especially those that respond to the environment (Antebi 2015). Several studies implicate steroid signaling in the PZ response to nutrients. DAF-12 is the best-characterized nuclear hormone receptor. In the dauer decision, DAF-12 acts downstream of the insulin and TGFβ signaling pathways (Fielenbach and Antebi 2008). DAF-12 also acts downstream of insulin-mediated larval PZ accumulation, but it does not act downstream of PEPT-1, TOR, or TGFβ (Hubbard et al. 2013), again indicating that the animal employs and integrates different signaling pathways in different ways depending on the physiological cue, the specific life stage, and the tissue-specific readout. In the dauer decision, the bile acid–like steroid hormone dafachronic acid (DA) binds DAF-12 to promote the reproductive fate and to deter dauer entry. Counterintuitively, in the adult, DA addition can interfere with germ cell proliferation in a daf-12–dependent manner (Mukherjee et al. 2017). A paralogous nuclear hormone receptor NHR-8 is also implicated PZ accumulation since nhr-8(rf) mutants are less sensitive to dietary restriction (Thondamal et al. 2015). However, nhr-8 does not appear to function downstream of DA in the tumor model that implicated daf-12 in the adult DA response (Mukherjee et al. 2017). Interestingly, like several other paralogous gene pairs that have different effects on or different temporal requirements in the germ line (e.g., Maciejowski et al. 2005; Agarwal et al. 2018), nhr-8 is located on an autosome while daf-12 is on the X chromosome.
Starvation arrest points
Overview of germline quiescence:
C. elegans employs a variety of mechanisms to reversibly arrest development in response to extremely poor environmental conditions. Unlike “low food” conditions described above that impair larval PZ accumulation, arrest occurs in response to total starvation or, in the case of dauer, persistent extreme conditions. Reversible arrest mechanisms delay or extend reproductive maturity and thereby allow the animal to coordinate reproduction with environmental conditions that are conducive to survival of their progeny. Perhaps one key to the evolutionary success of C. elegans is that in the face of extreme “boom and bust” resources (Frézal and Félix 2015) they can nevertheless produce some, if not many, progeny. Then, even in poor conditions, these progeny can themselves delay reproduction until conditions improve.
Although developmental arrest can occur upon starvation throughout development (Golden and Riddle 1984; Johnson et al. 1984; Schindler et al. 2014), the arrest points for which the germ line has been best characterized are L1 arrest (Baugh 2013), dauer (Fielenbach and Antebi 2008), and adult reproductive diapause (ARD) (Angelo and Van Gilst 2009; Seidel and Kimble 2011) (Figure 7C). These three environmentally induced arrest points differ in important ways with respect to germ cell quiescence. L1 arrest occurs upon total starvation at hatching. This arrest therefore prolongs the period of PGC quiescence that is established in the embryo prior to L1 starvation. Dauer arrest occurs over a prolonged time-frame of continuously poor conditions and involves radical morphological and metabolic changes in the animal. In contrast to L1 starvation, all germ cells are proliferative stem cells prior to dauer entry. Finally, ARD occurs when animals molt to adulthood after acute starvation in the L4. The germ line at this stage contains multiple germ cell stages including proliferating stem and progenitor cells, cells that have entered the meiotic pathway, and cells undergoing gametogenesis. The ARD state is characterized by loss of maturing germ cells and maintenance of a stem cell pool that can repopulate the germ line upon refeeding. The somatic backdrop at each arrest point is also different. Not surprisingly, the regulatory networks that govern germ cell quiescence for each of these conditions differ as well. We consider each of these germ cell arrest scenarios in turn.
L1:
When newly hatched L1 larvae emerge in the absence of food, they do not develop and can survive for weeks (Baugh 2013). Both somatic and germ cells of the L1 remain quiescent, although most somatic cells arrest in G1 while PGCs arrest in G2 (Hong et al. 1998; Fukuyama et al. 2006). The PGCs depend on daf-18 PTEN and aak AMPK (encoded by partially redundant aak-1 and aak-2) to remain quiescent in the continued absence of food, while somatic cells depend on DAF-16 FOXO (Baugh and Sternberg 2006; Fukuyama et al. 2006, 2012, 2015; Demoinet et al. 2017; Tenen and Greenwald 2019). This difference likely relates to the difference in cell cycle arrest point. Furthermore, overexpression of certain neuronal insulin-like ligands (ins-3, -4, or -9) (Zheng et al. 2018) causes germ cells to divide inappropriately in the absence of food. Thus, while developmental arrest of the soma and the germ line in starved L1 hatchlings are both under the control of insulin signaling, L1 somatic cell cycle arrest is under the control of daf-16 FOXO (Baugh and Sternberg 2006), while germ cell cycle arrest is dependent on daf-18 PTEN and aak AMPK (Fukuyama et al. 2006, 2012). In addition, certain somatic cells can be coaxed out of quiescence with dietary ethanol and amino acids, while PGCs are resistant (Fukuyama et al. 2015).
Depleting TORC1 pathway components partially suppresses the inappropriate proliferation of both daf-18 PTEN and aak AMPK mutant PGCs in starved L1 larvae, suggesting that TORC1 pathway components act downstream of daf-18 PTEN and aak AMPK in the regulation of PGCs in L1 starvation. The TOR pathway likely integrates food sensing, cellular starvation responses, and other downstream effects of daf-18 PTEN and aak AMPK (Fukuyama et al. 2012). However, daf-18 PTEN and aak AMPK do not act in a linear pathway since the PGC proliferation phenotype of the triple null mutant is more severe than the daf-18 PTEN single or aak-1/-2 AMPK double mutants alone (Demoinet et al. 2017).
Dauer:
Dauer is one of the best-understood animal diapause states (Fielenbach and Antebi 2008). The developmental decision to enter dauer (vs. continuing directly in the “reproductive” developmental mode) is made in the late L1 stage in response to temperature, overcrowding, and severe food limitation. Complex morphological and metabolic changes occur during the ensuing modified L2 stage (L2d), during which animals can still revert to a nondauer pathway if conditions improve (Riddle and Albert 1997). Although analysis of the germ cell arrest is complicated by the heterogeneity of germ cell cycle progression during the protracted time-frame of dauer entry, germline proliferation is progressively suppressed during the L2d (Narbonne and Roy 2006). If poor conditions persist, L2d animals enter the highly stress-resistant dauer stage (L3d) that is characterized by specific modifications of the pharynx, cuticle, and behavior. Worms can survive for months in dauer, far exceeding their normal life span (Klass and Hirsh 1976). Yet when conditions improve, animals exit dauer (as postdauer L3/L4) and become reproductive adults. Thus, during dauer, germline stem cells can remain viable for a long but uncertain duration.
Dauer-arrested germ cells are quiescent with respect to the cell cycle. In nondauer conditions, by the early L2 stage, all germ cells are mitotically competent, and they are capable of expressing genes associated with meiotic entry (Austin and Kimble 1987; Merritt and Seydoux 2010). However, upon dauer entry, germ cells arrest prior to the time at which nondauer germ cells would normally overtly enter the meiotic pathway. Consistent with a glp-1–dependent stem cell state, although they will eventually arrest in G2, dauer-destined germ cells (L2d) will prematurely enter meiosis in the absence of glp-1 (Narbonne and Roy 2006). This contrasts with experiments in the ARD scenario (see below).
Downstream of the insulin and TGFβ pathways, DAF-18 PTEN, AAK AMPK, and DAF-12 NHR are implicated in dauer germline arrest. Unlike aak AMPK, daf-18 PTEN is also required for somatic gonad blast cell arrest. Moreover, daf-18 PTEN is required in the somatic gonad blast cells to maintain quiescence of both the somatic gonad and the germ cells (Tenen and Greenwald 2019). The AMPK-activating kinase PAR-4 LKB1 also contributes to the maintenance of germ cell quiescence in dauer, although not solely through its effect on AMPK (Narbonne and Roy 2006). Remarkably, AMPK expression in neurons and the excretory system can restore postdauer fertility and dauer quiescence to aak mutants (Kadekar and Roy 2019). Consistent with its critical role in the dauer decision downstream of the insulin and DAF-7 dauer TGFβ signaling pathways, the DAF-12 NHR is implicated in germline stem cell arrest by way of the dauer-inducing complex it forms with DIN-1S when it is unliganded (Colella et al. 2016). Unlike DAF-18 PTEN and AAK AMPK, DIN-1S is required in the germ line for germ cell quiescence. Analysis of genetic interactions between these pathways suggests that both germline and somatic gonadal divisions are subject to parallel control by DIN-1S and AMPK, such that the double mutant causes an additive effect on inappropriate germ cell accumulation. In addition, a role for aak-2 in somatic gonad quiescence is revealed when din-1S activity is also reduced (Colella et al. 2016).
Although L1 arrest and dauer arrest use partially overlapping molecular pathways, these recent studies point to the existence of complex tissue-specific interactions that depend on the stage and the exact downstream outputs (e.g., germ cell arrest vs. effects on postarrest viability or fertility). The site of action of several relevant components remains to be determined, as does the nature of signal(s) from the soma to germ line.
Adult Reproductive Diapause (ARD):
Individual hermaphrodites that experience sudden acute starvation in the L4 stage meet one of three ends: they either arrest development just after the adult molt, suffer matricide due to hatching of embryos in the uterus, or exhibit ARD (Angelo and Van Gilst 2009) (Figure 7C). The exact proportions of animals in these groups depends on the activity of NHR-49, which is related to nuclear hormone receptors that are implicated in mammalian responses to food deprivation (Angelo and Van Gilst 2009), and on the exact time of starvation in the L4. Nevertheless, all animals are capable of entering ARD if embryonic development is arrested to prevent matricide (Seidel and Kimble 2011).
Adult hermaphrodite gonads in ARD contain a population of ∼35 distal undifferentiated germ cells in each gonad arm, abutting the DTC. The remainder of the germ line undergoes apoptosis and autophagy, resulting in a characteristically atrophied appearance (Angelo and Van Gilst 2009). During ARD, the germ cells appear to arrest in the G2, similar to other germ cell arrest points. However, as mentioned above, in contrast to dauer-destined germ cells, ARD arrested germ cells do not enter meiosis when glp-1 is depleted (Seidel and Kimble 2015). This difference may reflect the more gradual cessation of germline proliferation in dauer-inducing conditions relative to germ cells arrested in ARD, such that germ cells progressing to dauer can complete the terminal division required to enter meiosis, whereas arrested ARD germ cells cannot. Alternatively, it may reflect other regulatory differences. Remarkably, even after 30 days of ARD, the germ line can regrow and fertility can be restored when food is reintroduced (Angelo and Van Gilst 2009; Seidel and Kimble 2011). The initial rate of accumulation of germline stem cells during regrowth is similar to the pace of larval PZ accumulation, but the number of PZ cells does not reach level it would in continuously fed adults (Roy et al. 2016) (Figure 7C). Nevertheless, regrowth of the post-ARD PZ offers a convenient model to distinguish between developmentally regulated (i.e., larval vs. adult) responses to environmental fluctuation since regrowth after ARD expands the PZ during the adult stage (Roy et al. 2016). Somatic tissues of ARD-arrested and refed animals also undergo profound changes (Angelo and Van Gilst 2009). Restoration of the germ line upon refeeding is secondary to restoration of the soma (including intestinal appearance, pharyngeal pumping rate, and motility) and to optimal RNA metabolism (Burnaevskiy et al. 2018).
ARD enables survival without loss of fertility upon acute late-larval starvation. The life span of continuously fed laboratory animals is ∼2 weeks, while ARD animals can survive three times longer (Angelo and Van Gilst 2009). Although post-ARD self-fertility declines, likely due to loss of viable self-sperm, some animals can produce cross-progeny when mated after 30 days of ARD (Angelo and Van Gilst 2009). If similar conditions occur in the wild, starved L4 hermaphrodites that survive as ARD adults could presumably find food, mate, and reproduce after weeks of ARD.
Decline in the Germline Stem Cell System During Aging
While one focus of the aging field is how the germ line limits life span (Hsin and Kenyon 1999; Antebi 2012), here we discuss a different question: how does advanced adult age affect the PZ and germline stem cells? Given the conservation of cellular and molecular aspects of stem cell biology and of aging, studies of age-related changes in C. elegans germline stem cells have implications for aging stem cells in other organisms.
A general model is emerging whereby, similar to many mammalian stem cell systems, C. elegans germline stem cells are limited and become “depleted” with age. After a period of apparent homeostasis in early adulthood, the number of cells in the hermaphrodite PZ declines sharply starting from the third day of adulthood (Garigan et al. 2002; Killian and Hubbard 2005; Hughes et al. 2007; Luo et al. 2010; Shi and Murphy 2014; Narbonne et al. 2015; Qin and Hubbard 2015; Kocsisova et al. 2019) (Figure 7A). However, due to the peculiarities of C. elegans reproductive biology, this depletion can be moderated, possibly to optimize the opportunity for late-life mating and reproduction. Despite different endpoints and parameters measured, common observations indicate that PZ decline is not due to germ cell death, but rather to slower cell cycle progression and diminished GLP-1 Notch signaling (Narbonne et al. 2015; Qin and Hubbard 2015; Kocsisova et al. 2019). We first introduce salient features of the aging worm germ line, and then consider in turn, the adult PZ when sperm are depleted (with and without subsequent mating), aging in the continuous presence of sperm, and molecular aspects of these aging PZ phenotypes.
Aging progenitor zone in sperm-depleted hermaphrodites
In unmated C. elegans hermaphrodites, the cache of self-sperm produced in the L4 stage are largely consumed by day 3 of adulthood, as a result of continuous fertilization of available oocytes. Sperm depletion in the germ line in these adults or in genetically spermless hermaphrodites (females) substantially alters the biology of the worm, including mRNA levels of a largely overlapping set of ∼1000 genes (Angeles-Albores et al. 2017). With the decline in stored sperm, sperm-derived signals for oocyte maturation and ovulation decrease such that oocytes arrest in meiotic prophase and stack up in the proximal germ line (McCarter et al. 1999; Miller et al. 2001; Lee et al. 2007). In these animals, the pace of PZ decline and MI are both reduced relative to the conditions in which oocytes continuously mature and are ovulated. One possible explanation is that in the PZ of these adults, both mitotic cycle progression and the ability enter the meiotic pathway are slowed. Genetic analysis shows that these PZ changes are not due to loss of sperm signals or fertilization, but instead correlate with reduced flux of germ cells through the oogenic developmental assembly line (Narbonne et al. 2015; Qin and Hubbard 2015). When hermaphrodites are mated after self-sperm depletion, fewer cells are found in the PZ relative to unmated sperm-depleted hermaphrodites of the same age (Narbonne et al. 2015; Qin and Hubbard 2015; see below). These observations and others support a model in which the PZ is “used up” over time, but in which depletion occurs more slowly in unmated sperm-depleted hermaphrodites due to a reduced cell division rate and a likely slower rate of meiotic entry.
Aging progenitor zone in the continuous presence of sperm
When sperm are continuously provided, the PZ still becomes depleted over time and a marked decline in reproductive output occurs (Kocsisova et al. 2019). Importantly, the decline in progeny production largely occurs as a consequence of changes in the PZ, including (i) a reduced stem cell pool (inferred from a decreased extent of SYGL-1) and reduced GLP-1 signaling (inferred from a decreased extent of LST-1), (ii) overall slower cell cycle progression, and (iii) a reduced rate of meiotic entry. In addition, germ cells that do enter meiosis progress more slowly through prophase of meiosis I, leading to a decreased rate of oocyte production and decreased progeny production. Surprisingly, PZ changes are observed as early as adult day 3 (“early-midlife”), significantly earlier than major age-related changes in the soma and the age-related decline in viability. A somewhat comparable phenomenon occurs in human females, where loss of reproductive capacity begins in the third decade when the soma is essentially at peak function, and menopause occurs over a decade later. Could the decline in oocyte production in C. elegans be the worm analog of age-related reproductive decline and menopause in humans?
Unlike very old (adult day 8–12) adult worms, the gonads of day 3 adult worms are sufficiently durable to withstand the rigors of dissection and fixation required for immunohistochemistry, allowing a detailed cell cycle progression analysis that includes not only M and S phase index, but G2 duration (EdU time course combined with M phase), and duration of total non-S phases (Kocsisova et al. 2019). Unlike sperm-depleted hermaphrodites, slower cell cycle progression in the presence of sperm is not due to the cessation of oocyte flux since mated hermaphrodites continue to mature and ovulate oocytes. In addition, these relatively early changes are seen in virtually all animals in both gonad arms, suggesting the existence of systemic control mechanisms that differ from more local signals regulating the PZ in response to oocyte accumulation in the absence of sperm. The molecular mechanisms that affect the stem cell pool and that slow the cell cycle in sperm-replete hermaphrodites over time are unknown.
Early insights into molecular mechanisms
Genetic studies implicate known “aging” pathways in PZ decline, but with several twists. Mutations in the C. elegans IR daf-2 that confer extended life span also slow the decline of the PZ, in a manner dependent on daf-16 FOXO (Garigan et al. 2002; Luo et al. 2010; Qin and Hubbard 2015). However, this is unlikely to be due to a global antiaging signal from persistent DAF-16 FOXO activity in these mutants, since tissue requirements for daf-16(+) are mutually exclusive for life-span vs. germline PZ regulation (Libina et al. 2003; Qin and Hubbard 2015). Unlike the germline-autonomous PZ-limiting role for daf-16(+) during larval PZ expansion (Michaelson et al. 2010), the PZ-preserving activity of daf-16(+) in aged animals is required in a small number of proximal somatic gonad (PSG) cells (within the distal spermathecal and proximal uterine lineages). These cells do not directly contact the PZ, suggesting that an as-yet unidentified DAF-16 transcriptional target(s) in the PSG produces a product that influences the PZ during aging, either directly or indirectly. PSG–daf-16 also partially contributes to the regulation of PZ depletion by oocyte accumulation since the number of PZ cells in fog-2 (spermless) mutant hermaphrodites declines more quickly when daf-16 is depleted in the PSG, even though these hermaphrodites still accumulate arrested oocytes in the proximal germ line (Qin and Hubbard 2015). Reduced PZ MI in sperm-depleted hermaphrodites also requires daf-18 PTEN and AMPK pathway signaling, perhaps via accumulated oocytes within the same gonad arm (Narbonne et al. 2015, 2017).
Progenitor Zone Responses to the Environment and Aging: A Speculative Unifying Model
In line with a model of “optimal” rather than “maximal” progeny production (Hughes et al. 2007), we speculate that the signaling systems reporting to the PZ on food availability and competitor population density enable hermaphrodites to produce some, if not many, progeny in diverse environmental conditions. In this model, during larval stages, suboptimal conditions reduce PZ accumulation and reduce progeny production so as not to deplete dwindling resources, while severe conditions that lead to developmental arrest, especially dauer, enhance dispersal to potentially better environments while preserving fertility. During adulthood, strategies of ARD and slowed PZ loss upon sperm depletion prevent complete loss of the PZ and allow the possibility of late-life reproduction, should conditions improve and opportunities arise for sperm replenishment by mating.
Future Directions
Studies of C. elegans have provided significant insight into the regulation of stem cell systems in general, and for the germ line, how the stem cell system is modulated under diverse conditions to control reproductive output. Here, we highlight several areas for future study.
A large collection of genes that function in the stem cell vs. meiotic development decision (e.g., Table 2 and Table 3) remain to be fully characterized. As these genes only show a strong phenotype in a sensitized genetic background, they likely function redundantly with another gene or pathway. It is not currently known if these genes function primarily in fine-tuning the decision, have a major function under a specific condition, or represent key components that have not yet been realized because of redundancy. Additional genetic, molecular, and cellular analysis will shed light on their function. Furthermore, the connection between regulation of the mitotic cell cycle and stem cell fate is still mysterious.
Our current cellular understanding of the stem cell system is largely from single or multiple time-point studies of populations of worms. Longitudinal analyses of individual worms, ideally with long-term live imaging, will increase our understanding of the dynamics of the stem cell system over the course of development and the molecular and cellular transitions as germ cells move proximally and begin differentiation. Indeed, C. elegans is one of the few systems where it is possible to analyze germline stem cells in intact individuals over time. Complementary molecular approaches, such as cell lineage markers, may be possible in the near future. Long-term single-cell imaging is technically challenging because of the extreme sensitivity of C. elegans germline biology to mounting and imaging. New approaches and methodologies will likely mitigate these issues. Importantly, it will be necessary to demonstrate that any new experimental set up does not perturb the stem cell system.
The influence of the animal’s environment and of physiology on the stem cell system is another open area for which this stem cell system is a particularly tractable model. It will be important to identify sensory and metabolic cues that modulate the stem cell system and to identify the signaling mechanisms that convey physiologically relevant information to the stem cells and their progeny. These will likely vary depending on the animal’s life history and stage of development. Where cell cycle activity is regulated by environment and physiology, it will also be of interest to determine the exact molecular mechanism(s).
Despite advances in our understanding of mechanisms of germline fate, cell cycle regulation, and the consequences of physiological signaling, it is still unclear how these cellular mechanisms together lead to the observed emergent properties of the dynamic stem cell system at the tissue level. Individual cellular decisions and behavior, the animal’s life history, and subtleties of gonad morphology can all have unanticipated effects on tissue-level outcomes when considered in synergy over time. For example, the cell cycle rate of distal germ cells affects the rate of displacement of more proximal cells, which, in turn, affects cell fate decisions. Approaches that will facilitate a tissue-level understanding of the stem cell system are long-term live imaging of the entire gonad with relevant markers, in various genetic backgrounds, and under different environmental conditions. Integrating the behavior of single germline cells as they proliferate and differentiate over the course of development to generate the entire germ line remains an open challenge.
Acknowledgments
We thank Ariz Mohammad, Olga Pekar, David Hall, and Zuzana Kocsisova for supplying images. We also thank the reviewers for helpful comments. Related work was supported by the National Institutes of Health (grants R01GM61706 and R01GM102254 to E.J.A.H. and grant R01GM100756 to T.S.).
Appendix
Germ Cell Tumors and Other Germ Cell Abnormalities
Several different cellular and molecular mechanisms can give rise to germ cell tumors in C. elegans. Proliferative tumors can be broken down into two main categories: those that are dependent on GLP-1 Notch signaling and those are not. Teratomas represent a distinct tumor type. Defects in somatic gonad morphology can also give rise to tumorous germline phenotypes. We consider in an “Other, nontumor” category an additional germ cell abnormality associated with advanced age. Below, we propose a framework for tumor nomenclature. We provide defining characteristics, examples of each subtype, and, together with special features, we suggest practical ways to distinguish among tumors. Where relevant, we note additional special features.
The developmental, morphological, and signaling landscape of the C. elegans gonad causes confusion regarding germ cell tumors for many reasons. First, tumors of similar origin may cause several different phenotypes. For example, the gonads of different individuals carrying the same glp-1(gf) allele can display several different types of tumors at various penetrance depending on dosage (maternal and zygotic), growth conditions, and temperature. Second, the same individual gonad may change tumor type over time. For example, within a single gonad arm in an individual, an early adult proximal latent niche tumor may be pushed into the uterus by sheath cell contractions and pressure from more distal germ cells, only to appear normal and nontumorous for a time, and later display a late-onset tumor. Third, adult tumors of different types can appear morphologically similar, so tumors cannot be distinguished by their adult phenotypes alone. For example, proximal latent niche–dependent tumors and mitotic reentry tumors arise by different mechanisms but both result in adults with proliferating germ cells in the proximal gonad. Fourth, within the same gonad, phenotypes may not be mutually exclusive. For example, in gld-1 mutants, the same gonad displays phenotypes resulting from both a mitotic reentry tumor and teratoma. Fifth, genetic interactions can alter the penetrance of tumor formation or the size of tumors, or both. Finally, somatic gonad defects complicate interpretation of germline tumor phenotypes.
GLP-1 Notch pathway tumors
Full
Defining Characteristics: Germ cells fail to enter the meiotic pathway and continue to proliferate.
Example: Strong constitutively active allele of glp-1, glp-1(oz112) (Berry et al. 1997)
Special features: Tumors fill the gonad and often breach its boundaries. In older animals, tumors may fill the entire animal, killing it.
Late-onset
Defining Characteristics: Growth of tumor starting from the distal stem cell pool and continuing in a distal-to-proximal direction over time.
Example: glp-1(oz112)/glp-1(+) heterozygotes (Berry et al. 1997)
Special features: Animals bearing these tumors may be initially fertile, but later the gonad fills with germ cells, with the same consequences as the Full tumor.
Complex
Defining Characteristics: Similar to a “Full” or “Late-onset” tumor except that some cells have entered the meiotic pathway in a variable pattern among the proliferating germ cells, either scattered within the tumor or in discrete patches.
Example: glp-1(ar202) Class B, as described in (Pepper et al. 2003a).
Proximal, latent niche
Defining Characteristics: The presence of GLP-1-expressing germ cells, proximal to gametes (that is, between the spermatheca and gametes).
Examples: pro-1 loss-of-function (Killian and Hubbard 2004; McGovern et al. 2009).
Special features: This type of tumor occurs when the first onset of meiosis (“initial meiosis”, see text) is delayed and, as a result, GLP-1-responsive cells are inappropriately juxtaposed to neighboring proximal somatic cells that produce DSL ligands. In normal development, at the time when DSL-producing proximal cells are born, the germ cells that contact these cells have already differentiated and no longer express GLP-1 on their surface. Thus, the cells that form a Proximal latent niche-dependent tumor never entered meiosis, and these tumors will still form in the presence of mutations that cause meiotic arrest (e.g., certain mutations in the MAPK pathway; Lee et al. 2007). In this way, they differ from Mitotic re-entry tumors (see below). In addition, in most cases, these tumors can be prevented or, if they have already formed can be differentiated, by eliminating DSL ligand expression from the proximal somatic gonad (McGovern et al. 2009). A characteristic of adult Proximal latent niche-dependent tumors of slow-cycling larval origin (see Additional Special note 2, below) is that they are often accompanied by a PZ of reduced size relative to the stage-matched wild type.
-
Additional Special notes:
1. This cellular tumor-promoting mechanism was dubbed “latent niche” since the proximal somatic gonad cells that express DSL ligands have a “latent” capacity to act as a secondary stem cell niche (McGovern et al. 2009). In the wild-type situation, where germline development and somatic gonad development are developmentally matched, these proximal somatic cells would never express the capacity to act as a niche. The latent niche concept has relevance for human cancer. First, for cancer-promoting receptor activity such as Notch, if receptor-dependent tumor cells are inappropriately juxtaposed to DSL ligand-producing cells anywhere in the body, the ligand-producing cells could act as a latent niche and thereby support tumor growth, either of a primary tumor or in the context of a second-site metastasis. Second, an inappropriate cell-cell interaction alone could conceivably support tumor formation without further genetic change to the receptor gene in the tumor precursor cells. Third, the latent niche may partially account for the observation of tissue bias for metastasis of certain tumor types, as noted in the “seed and soil” hypothesis of Paget (Paget 1889; Langley and Fidler 2011).
2. Any delay in initial meiosis can contribute to the latent niche tumor, be it germline autonomous or non-autonomous in origin. Examples of a germline autonomous origins are certain glp-1(gf) mutants that retain ligand sensitivity (such as glp-1(ar202)) and therefore can respond inappropriately to the proximal ligand. An example of non-autonomous origin is loss of pro-1 function in gonadal sheath cells. Indeed, any defect that slows the proliferation of germ cells during larval stages can also contribute if it delays initial meiosis. This delay is often due to inadequate displacement of the DTC away from the proximal gonad, trapping slow-cycling germ cells in a GLP-1-responsive (nevertheless slow-cycling) mitotic state.
3. A counter-intuitive consequence of this scenario is that genetic or other perturbations that interfere with accumulation of the germ cell pool in early larval stages (e.g., due to slow germ cell proliferation) will enhance the penetrance of proximal tumor formation in glp-1(gf) mutant adults. These same perturbations may or may not interfere with the size (that is, the number of cells) of adult proximal tumors.
4. Proximal latent niche dependent tumors may become large and can also breach the gonad. However, in the presence of certain mutations that slow germ cell proliferation, they may be prevalent but small in cell number (Korta et al. 2012).
5. Latent niche ligands likely have other developmental or physiological roles. For example, APX-1, one of the DSL ligands expressed in the proximal gonad and that is responsible for the latent niche effect, is required for normal ovulation (McGovern et al. 2018).
Other tumors
Meiotic entry-defective tumors
Defining Characteristics: Germ cells fail to enter the meiotic pathway (defective differentiation) and continue to proliferate, even in the absence of GLP-1 Notch signaling.
Examples: gld-1 gld-2 double or gld-1 gld-2 glp-1 triple null mutant combinations (Kadyk and Kimble 1998).
Special features: Germ cells proliferate robustly and can break out of the gonad, killing the animal. These tumors underscore the fact that GLP-1 Notch activity is not required for germ cell proliferation per se.
Additional Special note: These tumors are not identical to glp-1(gf) tumors because they lack ectopic lst-1 and sygl-1 expression distant from any DSL ligand source.
Mitotic re-entry tumors
Defining Characteristics: Mitotic germ cells proximal to cells that have entered and progressed into meiosis.
Examples: loss of gld-1 or puf-8 (Francis et al. 1995a; Subramaniam and Seydoux 2003).
Special features: Tumors form from germ cells that previously entered the meiotic pathway. In gld-1 mutants, female germ cells progress to the pachytene stage of prophase of meiosis I and then re-enter the mitotic cell cycle. In puf-8 mutants, spermatogonia re-entered the mitotic cell cycle. In the case of gld-1, these tumors are due to aberrant expression of mitotic and/or other mRNAs that are normally repressed until later stages of germline development or embryogenesis.
-
Additional Special notes:
1. This type of tumor does not represent “de-differentiation” in the sense that these cells do not revert to a normal pre-meiotic state.
2. In the two examples cited above, the tumor phenotype is germline sex-specific. In this way, these tumors differ from Meiotic Entry-defective tumors and glp-1(gf) tumors.
3. Like Proximal latent niche-dependent tumors (see above), mitotic re-entry tumors can appear as a mass of mitotic germ cells in the proximal gonad. However, mitotic re-entry tumors form later in development since germ cells must first enter meiosis and then return to mitosis. Also, unlike Proximal latent niche-dependent tumors, mitotic re-entry tumors can be inhibited by meiotic arrest prior to the stage of meiosis at which mitotic re-entry occurs (see above, e.g. meiotic prophase arrest with MAPK mutants).
Teratoma
Defining Characteristics: Germ cells adopt somatic fates in situ, expressing somatic cell markers and morphological features.
Example: mex-3; gld-1 double mutant (Ciosk et al. 2006).
Special features: Teratomas form as a result of aberrant gene expression characteristic of somatic cells that is normally repressed in the germ line.
-
Additional Special note:
Although teratomas are referred to as “tumors”, the cells with somatic characteristics are a non-proliferating abnormality of germline origin. In the case of the mex-3; gld-1 double mutant, cells with somatic character are not proliferating, while adjacent germ cells can be proliferating.
Tumors caused by somatic gonad defects
Abnormal somatic gonad morphology or fate
Defining Characteristics: Germ cells proliferate robustly in combination with abnormal somatic gonad morphology or altered somatic cell fate specification.
Examples: hlh-12/mig-24 (Voutev and Hubbard 2008), lin-12 (Seydoux et al. 1990)
Special features: Somatic gonad morphological defects can cause inappropriate soma-germline contact or otherwise prolong or elevate exposure to DSL ligands that, in turn, prevent meiotic entry. This class includes mutants with early DTC migration defects (see Figure) as well as mutants such as in lin-12 (lf) that alter cell fate (not shown). Proximal germline tumors were first characterized in lin-12 mutants (Seydoux et al. 1990). It is also possible that germline tumor phenotypes accompanied by gross somatic gonad defects may be independent of DSL ligands.
Extra-gonadal germ cells
Defining Characteristics: Extra-gonadal germ cells in the body cavity and/or in contact with non-gonadal cells.
Example: epi-1 RNAi (Gordon et al. 2019).
Special features: This phenotype can occur as a result of early breakdown of the somatic gonad basement membrane.
-
Additional Special notes:
1. The stem cell status of extra-gonadal germ cells is yet to be established. If stem cells escape the gonad, they could conceivably respond to extra-gonadal DSL ligand expression analogous to the latent niche. The cell cycle status of these cells also remains to be established. Escaped germ cells may also respond to other currently undefined cues that promote a proliferative fate or that promote cell cycle progression.
2. It is possible that defects in the basement membrane surrounding the gonad originate in non-gonadal cells.
Other, non-tumor
Uterine endomitotic oocytes in old adults
Defining Characteristics: Large, sometimes degenerating, cells in the uterus bearing an enlarged nucleus.
Example: old adult worms (Wang et al. 2018).
Special features: In aged worms, oocytes infrequently undergo meiotic maturation and are ovulated into the uterus in the absence of sperm. In the uterus they can subsequently undergo endoreduplication. Degenerating embryos may be interspersed among endomitotic oocytes, complicating phenotypic analysis.
-
Additional Special note:
Although uterine endomitotic oocytes have been referred to as a “tumor”, they are a non-mitotically proliferating abnormality of germline origin.
Footnotes
Reference numbers for data available in public repositories and IRB numbers.
Communicating editor: G. Seydoux
Literature Cited
- Ables E. T., and Drummond-Barbosa D., 2013. Cyclin E controls Drosophila female germline stem cell maintenance independently of its role in proliferation by modulating responsiveness to niche signals. Development 140: 530–540. 10.1242/dev.088583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ables E. T., Laws K. M., and Drummond-Barbosa D., 2012. Control of adult stem cells in vivo by a dynamic physiological environment: diet-dependent systemic factors in Drosophila and beyond. Wiley Interdiscip. Rev. Dev. Biol. 1: 657–674. 10.1002/wdev.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal I., Farnow C., Jiang J., Kim K.-S., Leet D. E. et al. , 2018. HOP-1 presenilin deficiency causes a late-onset notch signaling phenotype that affects adult germline function in Caenorhabditis elegans. Genetics 208: 745–762. 10.1534/genetics.117.300605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M. V., Andrade-Navarro M. A., and Ketting R. F., 2019. Function and evolution of nematode RNAi pathways. Noncoding RNA 5: 8. 10.3390/ncrna5010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames K., Da Cunha D. S., Gonzalez B., Konta M., Lin F. et al. , 2017. A non-cell-autonomous role of BEC-1/BECN1/beclin1 in coordinating cell-cycle progression and stem cell proliferation during germline development. Curr. Biol. 27: 905–913. 10.1016/j.cub.2017.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini R., Goupil E., Labella S., Zetka M., Maddox A. S. et al. , 2014. C. elegans Anillin proteins regulate intercellular bridge stability and germline syncytial organization. J. Cell Biol. 206: 129–143 [Corrigenda: J. Cell Biol. 209: 467 (2015)]. 10.1083/jcb.201310117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles-Albores D., Leighton D. H. W., Tsou T., Khaw T. H., Antoshechkin I. et al. , 2017. The Caenorhabditis elegans female-like state: decoupling the transcriptomic effects of aging and sperm status. G3 (Bethesda) 7: 2969–2977. 10.1534/g3.117.300080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo G., and Van Gilst M. R., 2009. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Sci. (New York, NY) 326: 954–958. 10.1126/science.1178343 [DOI] [PubMed] [Google Scholar]
- Antebi A., Culotti J. G., and Hedgecock E. M., 1998. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development 125: 1191–1205. [DOI] [PubMed] [Google Scholar]
- Antebi A., 2012. Regulation of longevity by the reproductive system. Exp. Gerontol. 48: 1–7. 10.1016/j.exger.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi, A., 2015 Nuclear receptor signal transduction in C. elegans (June 9, 2015), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.64.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S., Ohmachi M., Berkseth M., Nayak S., Hansen D. et al. , 2011. MPK-1 ERK controls membrane organization in C. elegans oogenesis via a sex-determination module. Dev. Cell 20: 677–688. 10.1016/j.devcel.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell K., Qin Z., Gavaghan D., Kugler H., Hubbard E. J. A. et al. , 2015. Mechano-logical model of C. elegans germ line suggests feedback on the cell cycle. Development 142: 3902–3911. 10.1242/dev.126359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J., and Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. 10.1016/0092-8674(87)90128-0 [DOI] [PubMed] [Google Scholar]
- Austin M. U., Liau W.-S., Balamurugan K., Ashokkumar B., Said H. M. et al. , 2010. Knockout of the folate transporter folt-1 causes germline and somatic defects in C. elegans. BMC Dev. Biol. 10: 46 10.1186/1471-213X-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery, L., and Y. You, 2012 C. elegans feeding (May 21, 2012), WormBook, ed. The C. elegans Research Community WormBook, doi: 10.1895/wormbook.1.150.1, http://www.wormbook.org. [Google Scholar]
- Avgousti D. C., Palani S., Sherman Y., and Grishok A., 2012. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 31: 3821–3832. 10.1038/emboj.2012.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I, 2006. Chemosensation in C. elegans (October 25, 2006), WormBook, ed. The C. elegans Research Community WormBook., 10.1895/wormbook.1.123.1, http://www.wormbook.org. [DOI]
- Baugh L. R., 2013. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. 10.1534/genetics.113.150847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., and Sternberg P. W., 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16: 780–785. 10.1016/j.cub.2006.03.021 [DOI] [PubMed] [Google Scholar]
- Beanan M. J., and Strome S., 1992. Characterization of a germ-line proliferation mutation in C. elegans. Development 116: 755–766. [DOI] [PubMed] [Google Scholar]
- Belfiore M., Pugnale P., Saudan Z., and Puoti A., 2004. Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development 131: 2935–2945. 10.1242/dev.01154 [DOI] [PubMed] [Google Scholar]
- Bernstein D., Hook B., Hajarnavis A., Opperman L., and Wickens M., 2005. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA 11: 447–458. 10.1261/rna.7255805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry L. W., Westlund B., and Schedl T., 1997. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124: 925–936. [DOI] [PubMed] [Google Scholar]
- Bessler J. B., Reddy K. C., Hayashi M., Hodgkin J., and Villeneuve A. M., 2007. A role for Caenorhabditis elegans chromatin-associated protein HIM-17 in the proliferation vs. meiotic entry decision. Genetics 175: 2029–2037. 10.1534/genetics.107.070987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G. et al. , 2009. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell 17: 355–364. 10.1016/j.devcel.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., A. K. Sewell, Z. Wu, and M. Han, 2019 TOR signaling in Caenorhabditis elegans development, metabolism, and aging. Genetics 213: 329–360. [DOI] [PMC free article] [PubMed]
- Braeckman, B. P., K. Houthoofd, and J. R. Vanfleteren, 2009 Intermediary metabolism (February 16, 2009), WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.146.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J. L., and Schedl T., 2016. Germline stem cell differentiation entails regional control of cell fate regulator GLD-1 in Caenorhabditis elegans. Genetics 202: 1085–1103. 10.1534/genetics.115.185678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaevskiy N., Chen S., Mailig M., Reynolds A., Karanth S. et al. , 2018. Reactivation of RNA metabolism underlies somatic restoration after adult reproductive diapause in C. elegans. eLife 7: e36194 10.7554/eLife.36194.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butuči M., Williams A. B., Wong M. M., Kramer B., and Michael W. M., 2015. Zygotic genome activation triggers chromosome damage and checkpoint signaling in C. elegans primordial germ cells. Dev. Cell 34: 85–95. 10.1016/j.devcel.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Byrd D. T., Knobel K., Affeldt K., Crittenden S. L., and Kimble J., 2014. A DTC niche plexus surrounds the germline stem cell pool in Caenorhabditis elegans. PLoS One 9: e88372 10.1371/journal.pone.0088372.t001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çelen İ., Doh J. H., and Sabanayagam C. R., 2018. Effects of liquid cultivation on gene expression and phenotype of C. elegans. BMC Genomics 19: 562 10.1186/s12864-018-4948-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari S. N., Mukherjee M., Vagasi A. S., Bi G., Rahman M. M. et al. , 2016. Bacterial folates provide an exogenous signal for C. elegans germline stem cell proliferation. Dev. Cell 38: 33–46. 10.1016/j.devcel.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., and Greenwald I., 2004. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6: 183–192. 10.1016/S1534-5807(04)00021-8 [DOI] [PubMed] [Google Scholar]
- Chi C., Ronai D., Than M. T., Walker C. J., Sewell A. K. et al. , 2016. Nucleotide levels regulate germline proliferation through modulating GLP-1/Notch signaling in C. elegans. Genes Dev. 30: 307–320. 10.1101/gad.275107.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M. S., Yoo A. S., and Greenwald I., 2010. sel-11 and cdc-42, two negative modulators of LIN-12/Notch activity in C. elegans. PLoS One 5: e11885 10.1371/journal.pone.0011885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S., Kodoyianni V., Bosenberg M., Friedman L., and Kimble J., 1996. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H). Development 122: 1373–1383. [DOI] [PubMed] [Google Scholar]
- Cinalli R. M., and Lehmann R., 2013. A spindle-independent cleavage pathway controls germ cell formation in Drosophila. Nat. Cell Biol. 15: 839–845. 10.1038/ncb2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O., Crittenden S. L., Morgan D. E., and Kimble J., 2010. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc. Natl. Acad. Sci. USA 107: 2048–2053. 10.1073/pnas.0912704107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin A., Zheng L., Taylor P. H., Paz A., Zhang L. et al. , 2015. Semi-permeable diffusion barriers enhance patterning robustness in the C. elegans germline. Dev. Cell 35: 405–417. 10.1016/j.devcel.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., and Priess J. R., 2004. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 131: 4831–4841. 10.1242/dev.01352 [DOI] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., and Priess J. R., 2006. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Sci. (New York, NY) 311: 851–853. 10.1126/science.1122491 [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J. et al. , 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. 10.1016/j.cell.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella E., Li S., and Roy R., 2016. Developmental and cell cycle quiescence is mediated by the nuclear hormone receptor coregulator DIN-1S in the Caenorhabditis elegans dauer larva. Genetics 203: 1763–1776. 10.1534/genetics.116.191858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi A. K., Wightman B., and Chalfie M., 2015. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics 200: 387–407. 10.1534/genetics.115.176099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Troemel E. R., Evans T. C., and Kimble J., 1994. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120: 2901–2911. [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M. et al. , 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. 10.1038/nature754 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Leonhard K. A., Byrd D. T., and Kimble J., 2006. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17: 3051–3061. 10.1091/mbc.E06-03-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Seidel H. S., and Kimble J., 2017. Analysis of the C. elegans germline stem cell pool. Methods Mol. Biol. 1463: 1–33. 10.1007/978-1-4939-4017-2_1 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Lee C., Mohanty I., Battula S., Knobel K. et al. , 2019. Sexual dimorphism of niche architecture and regulation of the Caenorhabditis elegans germline stem cell pool. Mol. Biol. Cell 30: 1757–1769. 10.1091/mbc.E19-03-0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalfó D., Michaelson D., and Hubbard E. J. A., 2012. Sensory regulation of the C. elegans germline through TGF-beta-dependent signaling in the niche. Curr. Biol. 22: 712–719. 10.1016/j.cub.2012.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoinet E., Li S., and Roy R., 2017. AMPK blocks starvation-inducible transgenerational defects inCaenorhabditis elegans. Proc. Natl. Acad. Sci. USA 114: E2689–E2698. 10.1073/pnas.1616171114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., and Greenwald I., 2016. Determinants in the LIN-12/Notch intracellular domain that govern its activity and stability during Caenorhabditis elegans vulval development. G3 (Bethesda) 6: 3663–3670. 10.1534/g3.116.034363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett M., Westlund B., and Schedl T., 2009. METT-10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans. Genetics 183: 233–247. 10.1534/genetics.109.105270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T. G., Wen C., and Greenwald I., 2000. SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97: 7877–7881. 10.1073/pnas.97.14.7877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D., 2008. Stem cells, their niches and the systemic environment: an aging network. Genetics 180: 1787–1797. 10.1534/genetics.108.098244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann C. R., Kraemer B., Wickens M., and Kimble J., 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3: 697–710. 10.1016/S1534-5807(02)00322-2 [DOI] [PubMed] [Google Scholar]
- Eckmann C. R., Crittenden S. L., Suh N., and Kimble J., 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160. 10.1534/genetics.104.029264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. E., and Kimble J., 1995. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139: 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., and Swain P. S., 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186. 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Ezcurra M., 2018. Dissecting cause and effect in host-microbiome interactions using the combined worm-bug model system. Biogerontology 3: 1–12. 10.1007/s10522-018-9752-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., and Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22: 2149–2165. 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E. et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K., Wilkinson H. A., and Greenwald I., 1993. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development 119: 1019–1027. [DOI] [PubMed] [Google Scholar]
- Fox P. M., and Schedl T., 2015. Analysis of germline stem cell differentiation following loss of GLP-1 notch activity in Caenorhabditis elegans. Genetics 201: 167–184. 10.1534/genetics.115.178061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P. M., Vought V. E., Hanazawa M., Lee M.-H., Maine E. M. et al. , 2011. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138: 2223–2234. 10.1242/dev.059535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., and Schedl T., 1995a gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine E., and Schedl T., 1995b Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., McGrath G., Zhang J., Ruddy D. A., Sym M. et al. , 2002. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev. Cell 3: 85–97. 10.1016/S1534-5807(02)00189-2 [DOI] [PubMed] [Google Scholar]
- Frézal L., and Félix M.-A., 2015. C. elegans outside the Petri dish. eLife 4: e05849. 10.7554/eLife.05849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M., Rougvie A. E., and Rothman J. H., 2006. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16: 773–779. 10.1016/j.cub.2006.02.073 [DOI] [PubMed] [Google Scholar]
- Fukuyama M., Sakuma K., Park R., Kasuga H., Nagaya R. et al. , 2012. C. elegans AMPKs promote survival and arrest germline development during nutrient stress. Biol. Open 1: 929–936. 10.1242/bio.2012836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M., Kontani K., Katada T., and Rougvie A. E., 2015. The C. elegans hypodermis couples progenitor cell quiescence to the dietary state. Curr. Biol. 25: 1241–1248. 10.1016/j.cub.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Fuller M. T., and Spradling A. C., 2007. Male and female Drosophila germline stem cells: two versions of immortality. Sci. (New York, NY) 316: 402–404. 10.1126/science.1140861 [DOI] [PubMed] [Google Scholar]
- Furuta T., Joo H.-J., Trimmer K. A., Chen S.-Y., and Arur S., 2018. GSK-3 promotes S-phase entry and progression in C. elegans germline stem cells to maintain tissue output. Development 145: dev161042. 10.1242/dev.161042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D., Hsu A.-L., Fraser A. G., Kamath R. S., Ahringer J. et al. , 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161: 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold A. R., Ryan J., Vallée-Trudeau J.-N., Dorn J. F., Labbé J.-C. et al. , 2015. Investigating the regulation of stem and progenitor cell mitotic progression by in situ imaging. Curr. Biol. 25: 1123–1134. 10.1016/j.cub.2015.02.054 [DOI] [PubMed] [Google Scholar]
- Golden J. W., and Riddle D. L., 1984. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102: 368–378. 10.1016/0012-1606(84)90201-X [DOI] [PubMed] [Google Scholar]
- Gopal S., Boag P., and Pocock R., 2017. Automated three-dimensional reconstruction of the Caenorhabditis elegans germline. Dev. Biol. 432: 222–228. 10.1016/j.ydbio.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Gordon K. L., Payne S. G., Linden-High L. M., Pani A. M., Goldstein B. et al. , 2019. Ectopic germ cells can induce niche-like enwrapment by neighboring body wall muscle. Curr. Biol. 29: 823–833.e5. 10.1016/j.cub.2019.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C., Hepler W., Mickey K. M., and Priess J. R., 2000. aph-2 encodes a novel extracellular protein required for GLP-1-mediated signaling. Development 127: 2481–2492. [DOI] [PubMed] [Google Scholar]
- Goutte C., Tsunozaki M., Hale V. A., and Priess J. R., 2002. APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 99: 775–779. 10.1073/pnas.022523499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracida X., and Eckmann C. R., 2013. Fertility and germline stem cell maintenance under different diets requires nhr-114/HNF4 in C. elegans. Curr. Biol. 23: 607–613. 10.1016/j.cub.2013.02.034 [DOI] [PubMed] [Google Scholar]
- Greenwald I., 2012. Notch and the awesome power of genetics. Genetics 191: 655–669. 10.1534/genetics.112.141812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, I., and R. A. Kovall, 2013 Notch signaling: genetics and structure (January 17, 2013), WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.10.2, 10.1895/wormbook.1.10.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny, T. L., and C. Savage-Dunn, 2013 TGF-β signaling in C. elegans (July 10, 2013), WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.22.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny T. L., Lambie E., Hartwieg E., Horvitz H. R., and Hengartner M. O., 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Guo S., and Kemphues K. J., 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620. 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Gupta P., Leahul L., Wang X., Wang C., Bakos B. et al. , 2015. Proteasome regulation of the chromodomain protein MRG-1 controls the balance between proliferative fate and differentiation in the C. elegans germ line. Development 142: 291–302. 10.1242/dev.115147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnik S., Thomas Y., Guo Y., Stoecklin J., Neagu A. et al. , 2018. PRP-19, a conserved pre-mRNA processing factor and E3 ubiquitin ligase, inhibits the nuclear accumulation of GLP-1/Notch intracellular domain. Biol. Open 7: bio034066. 10.1242/bio.034066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T. et al. , 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. 10.1006/dbio.1999.9356 [DOI] [PubMed] [Google Scholar]
- Hall B. A., Piterman N., Hajnal A., and Fisher J., 2015. Emergent stem cell homeostasis in the C. elegans germline is revealed by hybrid modeling. Biophys. J. 109: 428–438. 10.1016/j.bpj.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., and Schedl T., 2006. The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line. Curr. Top. Dev. Biol. 76: 185–215. 10.1016/S0070-2153(06)76006-9 [DOI] [PubMed] [Google Scholar]
- Hansen D., and Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans, pp. 71–99 in Advances in experimental medicine and biology, Springer, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Hubbard E. J. A., and Schedl T., 2004a Multi-pathway control of the proliferation vs. meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 268: 342–357. 10.1016/j.ydbio.2003.12.023 [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., and Schedl T., 2004b Control of the proliferation vs. meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. 10.1242/dev.00916 [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Hall D. H., and Stern B. D., 1987. Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100: 365–382. [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., and Hall D. H., 1990. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron 4: 61–85. 10.1016/0896-6273(90)90444-K [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., and Kimble J., 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120: 2913–2924. [DOI] [PubMed] [Google Scholar]
- Hillers, K. J., V. Jantsch, E. Martinez-Perez, and J. L. Yanowitz, 2017 Meiosis (May 4, 2017), WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.178.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D., Oppenheim D., and Klass M., 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49: 200–219. 10.1016/0012-1606(76)90267-0 [DOI] [PubMed] [Google Scholar]
- Hong Y., Roy R., and Ambros V., 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125: 3585–3597. [DOI] [PubMed] [Google Scholar]
- Hsin H., and Kenyon C., 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362–366. 10.1038/20694 [DOI] [PubMed] [Google Scholar]
- Hsu H.-J., LaFever L., and Drummond-Barbosa D., 2008. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev. Biol. 313: 700–712. 10.1016/j.ydbio.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J. A., Wu G., Kitajewski J., and Greenwald I., 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11: 3182–3193. 10.1101/gad.11.23.3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J. A., Korta D. Z., and Dalfó D., 2013. Physiological control of germline development in Germ Cell Development in C. elegans, pp. 101–131 edited by Schedl T., Springer, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelgas-Morales G., and Greenstein D., 2018. Control of oocyte meiotic maturation in C. elegans. Semin. Cell Dev. Biol. 84: 90–99. 10.1016/j.semcdb.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Evason K., Xiong C., and Kornfeld K., 2007. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 3: e25 10.1371/journal.pgen.0030025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., and Seraphin B., 2008. EJCs at the heart of translational control. Cell 133: 213–216. 10.1016/j.cell.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Jan E., Motzny C. K., Graves L. E., and Goodwin E. B., 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18: 258–269. 10.1093/emboj/18.1.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V., Tang L., Pasierbek P., Penkner A., Nayak S. et al. , 2007. Caenorhabditis elegans prom-1 is required for meiotic prophase progression and homologous chromosome pairing. Mol. Biol. Cell 18: 4911–4920. 10.1091/mbc.e07-03-0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A. M., and Engebrecht J., 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308: 206–221. 10.1016/j.ydbio.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Jarriault S., and Greenwald I., 2005. Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev. Biol. 287: 1–10. 10.1016/j.ydbio.2005.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Verheyden J. M., and Kimble J., 2011. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet. 7: e1001348 10.1371/journal.pgen.1001348.g007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. E., Mitchell D. H., Kline S., Kemal R., and Foy J., 1984. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 28: 23–40. 10.1016/0047-6374(84)90150-7 [DOI] [PubMed] [Google Scholar]
- Jones A. R., and Schedl T., 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9: 1491–1504. 10.1101/gad.9.12.1491 [DOI] [PubMed] [Google Scholar]
- Jones A. R., Francis R., and Schedl T., 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165–183. 10.1006/dbio.1996.0293 [DOI] [PubMed] [Google Scholar]
- Jungkamp A.-C., Stoeckius M., Mecenas D., Grün D., Mastrobuoni G. et al. , 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 44: 828–840. 10.1016/j.molcel.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadekar P., and Roy R., 2019. AMPK regulates germline stem cell quiescence and integrity through an endogenous small RNA pathway. PLoS Biol. 17: e3000309 10.1371/journal.pbio.3000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., and Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kadyk L. C., Lambie E. J., and Kimble J., 1997. glp-3 is required for mitosis and meiosis in the Caenorhabditis elegans germ line. Genetics 145: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S.-H., Tseng C.-Y., Wan C.-L., Su Y.-H., Hsieh C.-C. et al. , 2015. Aging and insulin signaling differentially control normal and tumorous germline stem cells. Aging Cell 14: 25–34. 10.1111/acel.12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper B. D., Lamphear B. J., Deshpande A. M., Jankowska-Anyszka M., Aamodt E. J. et al. , 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275: 10590–10596. 10.1074/jbc.275.14.10590 [DOI] [PubMed] [Google Scholar]
- Kerins J. A., Hanazawa M., Dorsett M., and Schedl T., 2010. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation vs. meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev. Dyn. 239: 1555–1572. 10.1002/dvdy.22274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A. M., and Kimble J., 2010. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. USA 107: 3936–3941. 10.1073/pnas.1000495107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A., Crittenden S. L., Friend K., Sorensen E. B., Porter D. F. et al. , 2013. Germline stem cells and their regulation in the nematode Caenorhabditis elegans. Adv. Exp. Med. Biol. 786: 29–46. 10.1007/978-94-007-6621-1_3 [DOI] [PubMed] [Google Scholar]
- Kershner A. M., Shin H., Hansen T. J., and Kimble J., 2014. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc. Natl. Acad. Sci. USA 111: 3739–3744. 10.1073/pnas.1401861111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian D. J., and Hubbard E. J. A., 2004. C. elegans pro-1 activity is required for soma/germline interactions that influence proliferation and differentiation in the germ line. Development 131: 1267–1278. 10.1242/dev.01002 [DOI] [PubMed] [Google Scholar]
- Killian D. J., and Hubbard E. J. A., 2005. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev. Biol. 279: 322–335. 10.1016/j.ydbio.2004.12.021 [DOI] [PubMed] [Google Scholar]
- Kim Y., Kostow N., and Dernburg A. F., 2015. The chromosome Axis mediates feedback control of CHK-2 to ensure crossover formation in C. elegans. Dev. Cell 35: 247–261. 10.1016/j.devcel.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., T. Wilson, and Kimble J., 2010. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc Natl. Acad. Sci. USA. 107: 17445–17450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., and Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. 10.1016/0012-1606(79)90035-6 [DOI] [PubMed] [Google Scholar]
- Kimble J. E., and White J. G., 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81: 208–219. 10.1016/0012-1606(81)90284-0 [DOI] [PubMed] [Google Scholar]
- Kimble J., and Crittenden S. L., 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23: 405–433. 10.1146/annurev.cellbio.23.090506.123326 [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., Lander L. E., Wing J. P., He W. W., and Hedgecock E. M., 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85: 829–839. 10.1016/S0092-8674(00)81267-2 [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., Gohel S. P., and Hedgecock E. M., 2000. The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development 127: 5071–5082. [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., and van den Heuvel S., 2019. Developmental control of the cell cycle: insights from Caenorhabditis elegans. Genetics 211: 797–829. 10.1534/genetics.118.301643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass M., and Hirsh D., 1976. Non-ageing developmental variant of Caenorhabditis elegans. Nature 260: 523–525. 10.1038/260523a0 [DOI] [PubMed] [Google Scholar]
- Koch U., Lehal R., and Radtke F., 2013. Stem cells living with a Notch. Development 140: 689–704. 10.1242/dev.080614 [DOI] [PubMed] [Google Scholar]
- Kocsisova Z., Mohammad A., Kornfeld K., and Schedl T., 2018. Cell cycle analysis in the C. elegans germline with the thymidine analog EdU. J. Vis. Exp. 10.3791/58339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsisova Z., Kornfeld K., and Schedl T., 2019. Rapid population-wide declines in stem cell number and activity during reproductive aging in C. elegans. Development 146: dev173195. 10.1242/dev.173195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodoyianni V., Maine E. M., and Kimble J., 1992. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 3: 1199–1213. 10.1091/mbc.3.11.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H., Chao M. Y., Larkins-Ford J., Corkins M. E., Somers G. A. et al. , 2008. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 6: e196 10.1371/journal.pbio.0060196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., and Ilagan M. X. G., 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korta D. Z., Tuck S., and Hubbard E. J. A., 2012. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development 139: 859–870. 10.1242/dev.074047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R. A., Gebelein B., Sprinzak D., and Kopan R., 2017. The canonical notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell 41: 228–241. 10.1016/j.devcel.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B., Crittenden S., Gallegos M., Moulder G., Barstead R. et al. , 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009–1018. 10.1016/S0960-9822(99)80449-7 [DOI] [PubMed] [Google Scholar]
- Lam N., Chesney M. A., and Kimble J., 2006. Wnt signaling and CEH-22/tinman/Nkx2.5 specify a stem cell niche in C. elegans. Curr. Biol. 16: 287–295. 10.1016/j.cub.2005.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie E. J., and Kimble J., 1991. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112: 231–240. [DOI] [PubMed] [Google Scholar]
- Lamont L. B., Crittenden S. L., Bernstein D., Wickens M., and Kimble J., 2004. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7: 697–707. 10.1016/j.devcel.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Langley R. R., and Fidler I. J., 2011. The seed and soil hypothesis revisited-The role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 128: 2527–2535. 10.1002/ijc.26031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L. R., and Hansen M., 2012. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 23: 637–644. 10.1016/j.tem.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Moyle M. W., Budrewicz J., Mendoza-Lopez J., Oegema K. et al. , 2019. The G2-to-M transition is ensured by a dual mechanism that protects cyclin B from degradation by Cdc20-activated APC/C. Dev. Cell 10.1016/j.devcel.2019.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws K. M., and Drummond-Barbosa D., 2017. Control of Germline Stem Cell Lineages by Diet and Physiology, pp. 67–99 in Signaling-Mediated Control of Cell Division, Springer International Publishing, Cham. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H. H., and Schedl T., 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15: 2408–2420. 10.1101/gad.915901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., and Schedl T., 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18: 1047–1059. 10.1101/gad.1188404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., and Schedl T., 2010. C. elegans star proteins, GLD-1 and ASD-2, regulate specific RNA targets to control development. Adv. Exp. Med. Biol. 693: 106–122. 10.1007/978-1-4419-7005-3_8 [DOI] [PubMed] [Google Scholar]
- Lee M.-H., Ohmachi M., Arur S., Nayak S., Francis R. et al. , 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. 10.1534/genetics.107.081356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Sorensen E. B., Lynch T. R., and Kimble J., 2016. C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. eLife 5: e18370. 10.7554/eLife.18370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y. S., Lu T., and Seydoux G., 2017. Nanos promotes epigenetic reprograming of the germline by down-regulation of the THAP transcription factor LIN-15B. eLife 6: e30201. 10.7554/eLife.30201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R., 2012. Germline stem cells: origin and destiny. Stem Cells 10: 729–739. 10.1016/j.stem.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D., and Greenwald I., 1995. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature 377: 351–354. 10.1038/377351a0 [DOI] [PubMed] [Google Scholar]
- Levitan D., Yu G., St George Hyslop P., and Goutte C., 2001. APH-2/Nicastrin functions in LIN-12/notch signaling in the Caenorhabditis elegans somatic gonad. Dev. Biol. 240: 654–661. 10.1006/dbio.2001.0486 [DOI] [PubMed] [Google Scholar]
- Li X., and Greenwald I., 1997. HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc. Natl. Acad. Sci. USA 94: 12204–12209. 10.1073/pnas.94.22.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N., Berman J. R., and Kenyon C., 2003. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502. 10.1016/S0092-8674(03)00889-4 [DOI] [PubMed] [Google Scholar]
- Linden L. M., Gordon K. L., Pani A. M., Payne S. G., Garde A. et al. , 2017. Identification of regulators of germ stem cell enwrapment by its niche in C. elegans. Dev. Biol. 429: 271–284. 10.1016/j.ydbio.2017.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissemore J. L., Connors E., Liu Y., Qiao L., Yang B. et al. , 2018. The molecular chaperone HSP90 promotes notch signaling in the germline of Caenorhabditis elegans. G3 (Bethesda) 8: 1535–1544. 10.1534/g3.118.300551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., and Maine E. M., 2007. The Bro1-domain protein, EGO-2, promotes Notch signaling in Caenorhabditis elegans. Genetics 176: 2265–2277. 10.1534/genetics.107.071225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sato C., Cerletti M., and Wagers A., 2010. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 92: 367–409. 10.1016/S0070-2153(10)92012-7 [DOI] [PubMed] [Google Scholar]
- Losick V. P., Morris L. X., Fox D. T., and Spradling A., 2011. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev. Cell 21: 159–171. 10.1016/j.devcel.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Kleemann G. A., Ashraf J. M., Shaw W. M., and Murphy C. T., 2010. TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell 143: 299–312. 10.1016/j.cell.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald L. D., Knox A., and Hansen D., 2008. Proteasomal regulation of the proliferation vs. meiotic entry decision in the Caenorhabditis elegans germ line. Genetics 180: 905–920. 10.1534/genetics.108.091553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., Ahn J. H., Cipriani P. G., Killian D. J., Chaudhary A. L. et al. , 2005. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics 169: 1997–2011. 10.1534/genetics.104.040121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., Ugel N., Mishra B., Isopi M., and Hubbard E. J. A., 2006. Quantitative analysis of germline mitosis in adult C. elegans. Dev. Biol. 292: 142–151. 10.1016/j.ydbio.2005.12.046 [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., and Villeneuve A. M., 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15: 1674–1687. 10.1101/gad.902601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox A. S., Habermann B., Desai A., and Oegema K., 2005. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development 132: 2837–2848. 10.1242/dev.01828 [DOI] [PubMed] [Google Scholar]
- Maine E. M., Hansen D., Springer D., and Vought V. E., 2004. Caenorhabditis elegans atx-2 promotes germline proliferation and the oocyte fate. Genetics 168: 817–830. 10.1534/genetics.104.029355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S. E., Thorpe C. J., Martin P. R., Chamberlain S. H., and Bowerman B., 1994. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis elegans embryo. Development 120: 2305–2315. [DOI] [PubMed] [Google Scholar]
- Mantina P., Macdonald L., Kulaga A., Zhao L., and Hansen D., 2009. A mutation in teg-4, which encodes a protein homologous to the SAP130 pre-mRNA splicing factor, disrupts the balance between proliferation and differentiation in the C. elegans germ line. Mech. Dev. 126: 417–429. 10.1016/j.mod.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Marin V. A., and Evans T. C., 2003. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130: 2623–2632. 10.1242/dev.00486 [DOI] [PubMed] [Google Scholar]
- Matova N., and Cooley L., 2001. Comparative aspects of animal oogenesis. Dev. Biol. 231: 291–320. 10.1006/dbio.2000.0120 [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., and Schedl T., 1997. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181: 121–143. 10.1006/dbio.1996.8429 [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., and Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. 10.1006/dbio.1998.9109 [DOI] [PubMed] [Google Scholar]
- McGovern M., Voutev R., Maciejowski J., Corsi A. K., and Hubbard E. J. A., 2009. A “latent niche” mechanism for tumor initiation. Proc. Natl. Acad. Sci. USA 106: 11617–11622. 10.1073/pnas.0903768106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern M., Castaneda P. G., Pekar O., Vallier L. G., Cram E. J. et al. , 2018. The DSL ligand APX-1 is required for normal ovulation in C. elegans. Dev. Biol. 435: 162–169. 10.1016/j.ydbio.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., and Seydoux G., 2010. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development 137: 1787–1798. 10.1242/dev.050799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., and Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Korta D. Z., Capua Y., and Hubbard E. J. A., 2010. Insulin signaling promotes germline proliferation in C. elegans. Development 137: 671–680. 10.1242/dev.042523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M.-H., Kosinski M., Schedl T. et al. , 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Sci. (New York, NY) 291: 2144–2147. 10.1126/science.1057586 [DOI] [PubMed] [Google Scholar]
- Millonigg, S., R. Minasaki, M. Nousch, and C. R. Eckmann, 2014 GLD-4-mediated translational activation regulates the size of the proliferative germ cell pool in the adult C. elegans germ line. PLoS Genet. 10: e1004647. [Corrigenda: PLoS Genet. 12: e1005862 (2016)]. 10.1371/journal.pgen.1004647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Vanden Broek K., Wang C., Daryabeigi A., Jantsch V. et al. , 2018. Initiation of meiotic development is controlled by three post-transcriptional pathways in Caenorhabditis elegans. Genetics 209: 1197–1224. 10.1534/genetics.118.300985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S. J., and Kimble J., 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441: 1068–1074. 10.1038/nature04956 [DOI] [PubMed] [Google Scholar]
- Mukherjee M., Chaudhari S. N., Balachandran R. S., Vagasi A. S., and Kipreos E. T., 2017. Dafachronic acid inhibits C. elegans germ cell proliferation in a DAF-12-dependent manner. Dev. Biol. 432: 215–221. 10.1016/j.ydbio.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C. T., 2013 Insulin/insulin-like growth factor signaling in C. elegans (December 26, 2013), WormBook, edited by The C. elegans Research Community WormBook, 10.1895/wormbook.1.164.1, http://www.wormbook.org. [DOI] [Google Scholar]
- Nadarajan S., Govindan J. A., McGovern M., Hubbard E. J. A., and Greenstein D., 2009. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 136: 2223–2234. 10.1242/dev.034603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., and Frøkjær-Jensen C., 2019. The Caenorhabditis elegans transgenic toolbox. Genetics 212: 959–990. 10.1534/genetics.119.301506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., and Roy R., 2006. Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development 133: 611–619. 10.1242/dev.02232 [DOI] [PubMed] [Google Scholar]
- Narbonne P., Maddox P. S., and Labbé J.-C., 2015. DAF-18/PTEN locally antagonizes insulin signalling to couple germline stem cell proliferation to oocyte needs in C. elegans. Development 142: 4230–4241. 10.1242/dev.130252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., Maddox P. S., and Labbé J.-C., 2017. DAF-18/PTEN signals through AAK-1/AMPK to inhibit MPK-1/MAPK in feedback control of germline stem cell proliferation. PLoS Genet. 13: e1006738 10.1371/journal.pgen.1006738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S., Santiago F. E., Jin H., Lin D., Schedl T. et al. , 2002. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12: 277–287. 10.1016/S0960-9822(02)00682-6 [DOI] [PubMed] [Google Scholar]
- Nousch M., Yeroslaviz A., Habermann B., and Eckmann C. R., 2014. The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms. Nucleic Acids Res. 42: 11622–11633. 10.1093/nar/gku838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M., Minasaki R., and Eckmann C. R., 2017. Polyadenylation is the key aspect of GLD-2 function in C. elegans. RNA 23: 1180–1187. 10.1261/rna.061473.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M. P., Chao P.-H., Kammenga J. E., and Sengupta P., 2018. Rictor/TORC2 mediates gut-to-brain signaling in the regulation of phenotypic plasticity in C. elegans. PLoS Genet. 14: e1007213 10.1371/journal.pgen.1007213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Lee Y. D., and Wagers A. J., 2014. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat. Med. 20: 870–880. 10.1038/nm.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki L., and Brand A. H., 2018. Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Sci. (New York, NY) 360: 99–102. 10.1126/science.aan8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget S., 1889. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 8: 98–101. [PubMed] [Google Scholar]
- Pekar O., Ow M. C., Hui K. Y., Noyes M. B., Hall S. E. et al. , 2017. Linking the environment, DAF-7/TGFβ signaling and LAG-2/DSL ligand expression in the germline stem cell niche. Development 144: 2896–2906. 10.1242/dev.147660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A. M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T. et al. , 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139: 920–933. 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Pepper A. S.-R., Killian D. J., and Hubbard E. J. A., 2003a Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A. S.-R., Lo T. W., Killian D. J., Hall D. H., and Hubbard E. J. A., 2003b The establishment of Caenorhabditis elegans germline pattern is controlled by overlapping proximal and distal somatic gonad signals. Dev. Biol. 259: 336–350. 10.1016/S0012-1606(03)00203-3 [DOI] [PubMed] [Google Scholar]
- Petcherski A. G., and Kimble J., 2000. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 405: 364–368. 10.1038/35012645 [DOI] [PubMed] [Google Scholar]
- Priess, J. R., 2005 Notch signaling in the C. elegans embryo (June 25, 2005), WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.4.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A., and Kimble J., 1999. The Caenorhabditis elegans sex determination gene mog-1 encodes a member of the DEAH-Box protein family. Mol. Cell. Biol. 19: 2189–2197. 10.1128/MCB.19.3.2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A., and Kimble J., 2000. The hermaphrodite sperm/oocyte switch requires the Caenorhabditis elegans homologs of PRP2 and PRP22. Proc. Natl. Acad. Sci. USA 97: 3276–3281. 10.1073/pnas.97.7.3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L., Lissemore J. L., Shu P., Smardon A., Gelber M. B. et al. , 1995. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics 141: 551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z., and Hubbard E. J. A., 2015. Non-autonomous DAF-16/FOXO activity antagonizes age-related loss of C. elegans germline stem/progenitor cells. Nat. Commun. 6: 7107 10.1038/ncomms8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racher H., and Hansen D., 2012. PUF-8, a Pumilio homolog, inhibits the proliferative fate in the Caenorhabditis elegans germline. G3 (Bethesda) 2: 1197–1205. 10.1534/g3.112.003350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiders S. A., Eastwood M. D., Bacher M., and Priess J. R., 2018. Binucleate germ cells in Caenorhabditis elegans are removed by physiological apoptosis. PLoS Genet. 14: e1007417 10.1371/journal.pgen.1007417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Peskin C. S., Tranchina D., Vargas D. Y., and Tyagi S., 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4: e309 10.1371/journal.pbio.0040309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S., Borgo B., Pazdernik N., Fox P., Mardis E. R. et al. , 2015. Caenorhabditis elegans glp-4 encodes a valyl aminoacyl tRNA synthetase. G3 (Bethesda) 5: 2719–2728. 10.1534/g3.115.021899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratliff M., Hill-Harfe K. L., Gleason E. J., Ling H., Kroft T. L. et al. , 2018. MIB-1 is required for spermatogenesis and facilitates LIN-12 and GLP-1 activity in Caenorhabditis elegans. Genetics 209: 173–193. 10.1534/genetics.118.300807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P., Lim C. S., Johnsen R., Albert P. S., Pilgrim D. et al. , 1996. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274: 1389–1391. 10.1126/science.274.5291.1389 [DOI] [PubMed] [Google Scholar]
- Rhoades J. L., Nelson J. C., Nwabudike I., Yu S. K., McLachlan I. G. et al. , 2019. ASICs mediate food responses in an enteric serotonergic neuron that controls foraging behaviors. Cell 176: 85–97.e14. 10.1016/j.cell.2018.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., and Albert P. S., 1997. Genetic and Environmental Regulation of Dauer Larva Development, pp. 739–768 in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., and Priess J. R., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Rosu S., and Cohen-Fix O., 2017. Live-imaging analysis of germ cell proliferation in the C. elegans adult supports a stochastic model for stem cell proliferation. Dev. Biol. 423: 93–100. 10.1016/j.ydbio.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Michaelson D., Hochman T., Santella A., Bao Z. et al. , 2016. Cell cycle features of C. elegans germline stem/progenitor cells vary temporally and spatially. Dev. Biol. 409: 261–271. 10.1016/j.ydbio.2015.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Kahler D. J., Yun C., and Hubbard E. J. A., 2018. Functional interactions between rsks-1/S6K, glp-1/Notch, and regulators of Caenorhabditis elegans fertility and germline stem cell maintenance. G3 (Bethesda) 8: 3293–3309. 10.1534/g3.118.200511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S., and van den Heuvel S., 2015. G1/S inhibitors and the SWI/SNF complex control cell-cycle exit during muscle differentiation. Cell 162: 300–313. 10.1016/j.cell.2015.06.013 [DOI] [PubMed] [Google Scholar]
- Rybarska A., Harterink M., Jedamzik B., Kupinski A. P., Schmid M. et al. , 2009. GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS Genet. 5: e1000494 10.1371/journal.pgen.1000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar K., Gu A., Xu X., Au V., Taylor J. et al. , 2016. UBR-5, a conserved HECT-type E3 ubiquitin ligase, negatively regulates Notch-type signaling in Caenorhabditis elegans. G3 (Bethesda) 6: 2125–2134. 10.1534/g3.116.027805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R. A., and Sabatini D. M., 2017. mTOR signaling in growth, metabolism, and disease. Cell 168: 960–976 (erratum: Cell 169: 361–371). 10.1016/j.cell.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., and Thomas J. H., 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17: 719–728. 10.1016/S0896-6273(00)80203-2 [DOI] [PubMed] [Google Scholar]
- Schaner C. E., Deshpande G., Schedl P. D., and Kelly W. G., 2003. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5: 747–757. 10.1016/S1534-5807(03)00327-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler A. J., Baugh L. R., and Sherwood D. R., 2014. Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet. 10: e1004426 10.1371/journal.pgen.1004426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Kuchler B., and Eckmann C. R., 2009. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 23: 824–836. 10.1101/gad.494009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R., 1978. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4: 7–25. [PubMed] [Google Scholar]
- Schulenburg H., and Félix M.-A., 2017. The Natural Biotic Environment of Caenorhabditis elegans. Genetics 206: 55–86. 10.1534/genetics.116.195511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. B., and Sinclair D. A., 2016. When stem cells grow old: phenotypes and mechanisms of stem cell aging. Development 143: 3–14. 10.1242/dev.130633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., and Kimble J., 2011. The oogenic germline starvation response in C. elegans. PLoS One 6: e28074 10.1371/journal.pone.0028074.g005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., and Kimble J., 2015. Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. eLife 4: e10832. 10.7554/eLife.10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Smith T. A., Evans J. K., Stamper J. Q., Mast T. G. et al. , 2018. C. elegans germ cells divide and differentiate in a folded tissue. Dev. Biol. 442: 173–187. 10.1016/j.ydbio.2018.07.013 [DOI] [PubMed] [Google Scholar]
- Severson A. F., and Meyer B. J., 2014. Divergent kleisin subunits of cohesin specify mechanisms to tether and release meiotic chromosomes. eLife 3: e03467 10.7554/eLife.03467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Schedl T., and Greenwald I., 1990. Cell-cell interactions prevent a potential inductive interaction between soma and germ line in C. elegans. Cell 61: 939–951. 10.1016/0092-8674(90)90060-R [DOI] [PubMed] [Google Scholar]
- Seydoux G., 2018. The P granules of C. elegans: A genetic model for the study of RNA–protein condensates. J. Mol. Biol. 430: 4702–4710. 10.1016/j.jmb.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Xu X., Fedotov A., Kelly W. G., and Maine E. M., 2009. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 5: e1000624 10.1371/journal.pgen.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., and Murphy C. T., 2014. Mating induces shrinking and death in Caenorhabditis mothers. Sci. (New York, NY) 343: 536–540. 10.1126/science.1242958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Haupt K. A., Kershner A. M., Kroll-Conner P., Wickens M. et al. , 2017. SYGL-1 and LST-1 link niche signaling to PUF RNA repression for stem cell maintenance in Caenorhabditis elegans. PLoS Genet. 13: e1007121 10.1371/journal.pgen.1007121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons B. D., and Clevers H., 2011. Strategies for Homeostatic Stem Cell Self-Renewal in Adult Tissues. Cell 145: 851–862. 10.1016/j.cell.2011.05.033 [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke J. M., Stacey S. C., Klein M. E., Mackin N. et al. , 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10: 169–178. 10.1016/S0960-9822(00)00323-7 [DOI] [PubMed] [Google Scholar]
- Spanier B., Wallwitz J., Zapoglou D., Idrissou B. M. G., Fischer C. et al. , 2018. The reproduction rate of peptide transporter PEPT-1 deficient C. elegans is dependent on dietary glutamate supply. Front. Mol. Biosci. 5: 109 10.3389/fmolb.2018.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Huelgas-Morales G., Tsukamoto T., Greenstein D., 2018. Multiple mechanisms inactivate the LIN-41 RNA-binding protein to ensure a robust oocyte-to-embryo transition in Caenorhabditis elegans. Genetics 210: 1011-1037. 10.1534/genetics.118.301421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A., Fuller M. T., Braun R. E., and Yoshida S., 2011. Germline stem cells. Cold Spring Harb. Perspect. Biol. 3: a002642 10.1101/cshperspect.a002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich T. A., Hall D. H., and Greenstein D., 2014. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 198: 1127–1153. 10.1534/genetics.114.168815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., and Updike D., 2015. Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 16: 406–416. 10.1038/nrm4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T. T., Campbell S. D., and O’Farrell P. H., 1998. The cell cycle program in germ cells of the Drosophila embryo. Dev. Biol. 196: 160–170. 10.1006/dbio.1998.8855 [DOI] [PubMed] [Google Scholar]
- Subramaniam K., and Seydoux G., 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871. [DOI] [PubMed] [Google Scholar]
- Subramaniam K., and Seydoux G., 2003. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr. Biol. 13: 134–139. 10.1016/S0960-9822(03)00005-8 [DOI] [PubMed] [Google Scholar]
- Suh N., Jedamzik B., Eckmann C. R., Wickens M., and Kimble J., 2006. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. USA 103: 15108–15112. 10.1073/pnas.0607050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N., Crittenden S. L., Goldstrohm A., Hook B., Thompson B. et al. , 2009. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181: 1249–1260. 10.1534/genetics.108.099440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., and Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., and Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Swiatek P., Kubrakiewicz J., and Klag J., 2009. Formation of germ-line cysts with a central cytoplasmic core is accompanied by specific orientation of mitotic spindles and partitioning of existing intercellular bridges. Cell Tissue Res. 337: 137–148. 10.1007/s00441-009-0788-8 [DOI] [PubMed] [Google Scholar]
- Tax F. E., Yeargers J. J., and Thomas J. H., 1994. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with delta and serrate of Drosophila. Nature 368: 150–154. 10.1038/368150a0 [DOI] [PubMed] [Google Scholar]
- Tax F. E., Thomas J. H., Ferguson E. L., and Horvitz H. R., 1997. Identification and characterization of genes that interact with lin-12 in Caenorhabditis elegans. Genetics 147: 1675–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen C. C., and Greenwald I., 2019. Cell non-autonomous function of daf-18/PTEN in the somatic gonad coordinates somatic gonad and germline development in C. elegans dauer larvae. Curr. Biol. 29: 1064–1072.e8. 10.1016/j.cub.2019.01.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil K., Herzog M., and Rajewsky N., 2018. Post-transcriptional Regulation by 3′ UTRs Can Be Masked by Regulatory Elements in 5′ UTRs. Cell Reports 22: 3217–3226. 10.1016/j.celrep.2018.02.094 [DOI] [PubMed] [Google Scholar]
- Thompson B. E., Bernstein D. S., Bachorik J. L., Petcherski A. G., Wickens M. et al. , 2005. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development 132: 3471–3481. 10.1242/dev.01921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thondamal M., Witting M., Schmitt-Kopplin P., and Aguilaniu H., 2015. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat. Commun. 5: 1–8. 10.1038/ncomms5879 [DOI] [PubMed] [Google Scholar]
- Tian X., Hansen D., Schedl T., and Skeath J. B., 2004. Epsin potentiates Notch pathway activity in Drosophila and C. elegans. Development 131: 5807–5815. 10.1242/dev.01459 [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L., and Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Updike D. L., and Strome S., 2009. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics 183: 1397–1419. 10.1534/genetics.109.110171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid S., Ariz M., Chaturbedi A., Anil Kumar G., and Subramaniam K., 2013. PUF-8 negatively regulates RAS/MAPK signalling to promote differentiation of C. elegans germ cells. Development 140: 1645–1654. 10.1242/dev.088013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijnberk L. M., van der Horst S. E. M., van den Heuvel S., and Ruijtenberg S., 2017. A dual transcriptional reporter and CDK-activity sensor marks cell cycle entry and progression in C. elegans. PLoS One 12: e0171600 10.1371/journal.pone.0171600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tetering G., van Diest P., Verlaan I., van der Wall E., Kopan R. et al. , 2009. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284: 31018–31027. 10.1074/jbc.M109.006775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M., Schroeter E. H., Pan Y., Blandford M., and Kopan R., 2004. Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J. Biol. Chem. 279: 50864–50873. 10.1074/jbc.M409430200 [DOI] [PubMed] [Google Scholar]
- Voronina E., Paix A., and Seydoux G., 2012. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 139: 3732–3740. 10.1242/dev.083980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vought V. E., Ohmachi M., Lee M.-H., and Maine E. M., 2005. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics 170: 1121–1132. 10.1534/genetics.105.042135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R., and Hubbard E. J. A., 2008. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics 180: 103–119. 10.1534/genetics.108.090274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R., Killian D. J., Ahn J. H., and Hubbard E. J. A., 2006. Alterations in ribosome biogenesis cause specific defects in C. elegans hermaphrodite gonadogenesis. Dev. Biol. 298: 45–58. 10.1016/j.ydbio.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Wang J. T., and Seydoux G., 2013. Germ cell specification. Adv. Exp. Med. Biol. 757: 17–39. 10.1007/978-1-4614-4015-4_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M., and Kimble J., 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. 10.1038/nature01039 [DOI] [PubMed] [Google Scholar]
- Wang C., Wilson-Berry L., Schedl T., and Hansen D., 2012. TEG-1 CD2BP2 regulates stem cell proliferation and sex determination in the C. elegans germ line and physically interacts with the UAF-1 U2AF65 splicing factor. Dev. Dyn. 241: 505–521. 10.1002/dvdy.23735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gupta P., Fairbanks J., and Hansen D., 2014. Protein kinase CK2 both promotes robust proliferation and inhibits the proliferative fate in the C. elegans germ line. Dev. Biol. 392: 26–41. 10.1016/j.ydbio.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhao Y., Ezcurra M., Benedetto A., Gilliat A. F., et al. , 2018. A parthenogenetic quasi-program causes teratoma-like tumors during aging in wild-type C. elegans. npj Aging Mech. Dis. 10.1038/s41514-018-0025-3 [DOI] [PMC free article] [PubMed]
- Watts J. L., and Ristow M., 2017. Lipid and Carbohydrate Metabolism in Caenorhabditis elegans. Genetics 207: 413–446. 10.1534/genetics.117.300106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Metzstein M. M., and Greenwald I., 1997. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development 124: 4759–4767. [DOI] [PubMed] [Google Scholar]
- Weng A. P., Ferrando A. A., Lee W., Morris J. P., Silverman L. B. et al. , 2004. Activating Mutations of NOTCH1 in Human T Cell Acute Lymphoblastic Leukemia. Sci. (New York, NY) 306: 269–271. 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Westlund B., Parry D., Clover R., Basson M., and Johnson C. D., 1999. Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1 in Notch-pathway signaling. Proc. Natl. Acad. Sci. USA 96: 2497–2502. 10.1073/pnas.96.5.2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Bernstein D. S., Kimble J., and Parker R., 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18: 150–157. 10.1016/S0168-9525(01)02616-6 [DOI] [PubMed] [Google Scholar]
- Wilkinson H. A., Fitzgerald K., and Greenwald I., 1994. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79: 1187–1198. 10.1016/0092-8674(94)90010-8 [DOI] [PubMed] [Google Scholar]
- Witham E., Comunian C., Ratanpal H., Skora S., Zimmer M. et al. , 2016. C. elegans Body Cavity Neurons Are Homeostatic Sensors that Integrate Fluctuations in Oxygen Availability and Internal Nutrient Reserves. Cell Reports 14: 1641–1654. 10.1016/j.celrep.2016.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke U., Jezuit E. A., and Priess J. R., 2007. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 134: 2227–2236. 10.1242/dev.004952 [DOI] [PubMed] [Google Scholar]
- Wong M. M., Belew M. D., Kwieraga A., Nhan J. D., and Michael W. M., 2018. Programmed DNA Breaks Activate the Germline Genome in Caenorhabditis elegans. Dev. Cell 46: 302–315.e5. 10.1016/j.devcel.2018.07.002 [DOI] [PubMed] [Google Scholar]
- Wright J. E., Gaidatzis D., Senften M., Farley B. M., Westhof E. et al. , 2011. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J 30: 533–545. 10.1038/emboj.2010.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Hubbard E. J., Kitajewski J. K., Greenwald I., 1998. Evidence for functional and physical association between Caenorhabditis elegans SEL-10, a Cdc4p-related protein, and SEL-12 presenilin. Proc. Natl. Acad. Sci. USA 95: 15787–15791. 10.1073/pnas.95.26.15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C.-C. G. et al. , 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757. 10.1016/j.cell.2006.09.033 [DOI] [PubMed] [Google Scholar]
- Yochem J., and Greenwald I., 1989. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell 58: 553–563. 10.1016/0092-8674(89)90436-4 [DOI] [PubMed] [Google Scholar]
- Yoo A. S., Bais C., and Greenwald I., 2004. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Sci. (New York, NY) 303: 663–666. 10.1126/science.1091639 [DOI] [PubMed] [Google Scholar]
- Yu X., Vought V. E., Conradt B., and Maine E. M., 2006. Eukaryotic translation initiation factor 5B activity regulates larval growth rate and germline development in Caenorhabditis elegans. Genesis 44: 412–418. 10.1002/dvg.20232 [DOI] [PubMed] [Google Scholar]
- Zanetti S., and Puoti A., 2013. Sex determination in the Caenorhabditis elegans germline. Adv. Exp. Med. Biol. 757: 41–69. 10.1007/978-1-4614-4015-4_3 [DOI] [PubMed] [Google Scholar]
- Zanetti S., Meola M., Bochud A., and Puoti A., 2011. Role of the C. elegans U2 snRNP protein MOG-2 in sex determination, meiosis, and splice site selection. Dev. Biol. 354: 232–241. 10.1016/j.ydbio.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Zetka M. C., Kawasaki I., Strome S., and Müller F., 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13: 2258–2270. 10.1101/gad.13.17.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S. et al. , 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. 10.1038/37297 [DOI] [PubMed] [Google Scholar]
- Zhang J., Holdorf A. D., and Walhout A. J. M., 2017. C. elegans and its bacterial diet as a model for systems-level understanding of host–microbiota interactions. Curr. Opin. Biotechnol. 46: 74–80. 10.1016/j.copbio.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Chiu H., Boudreau J., Papanicolaou T., Bendena W., et al. , 2018. A functional study of all 40 Caenorhabditis elegans insulin-like peptides. J. Biol. Chem. 293: 16912–16922. 10.1074/jbc.RA118.004542 [DOI] [PMC free article] [PubMed] [Google Scholar]