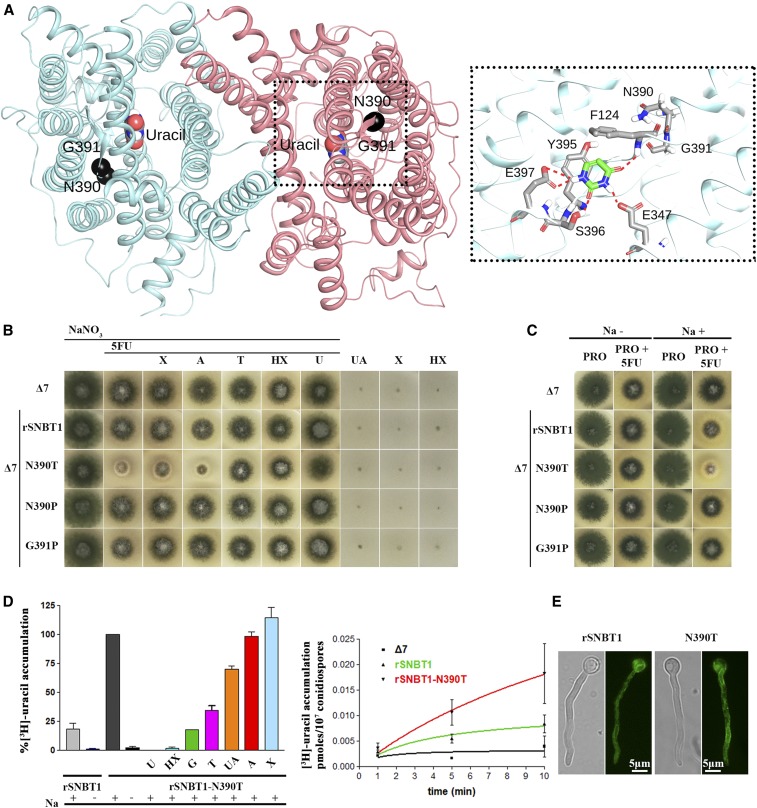
Figure 7.
Manipulation of a residue topologically equivalent to T401P leads to functional expression of a mammalian NAT homolog in A. nidulans. (A) Homology modeling of the topology of rSNBT1, constructed using, as described in the Materials and Methods, the inward-facing conformation of the crystal structure of the UapA dimer. The two mutated and functionally analyzed residues, N390 and G391, are shown as black spheres. The location of uracil, the major substrate of rSNBT1, is also depicted, as determined by dynamic docking (left panel). In the right panel, a zoomed-out picture of the substrate-binding site depicting the major interactions of uracil with specific residues. (B) Growth tests of isogenic A. nidulans strains expressing single-copy wild-type rSNBT1 or its mutated versions rSNBT1-N390T, rSNBT1-N390P, and rSNBT1-G391P. A negative control strain (i.e., the recipient Δ7 strain that has null activity for nucleobase transport; see text) is included for comparison. Growth tests were performed at 37° on minimal media supplemented with Na+ (100 mM NaCl); 10 mM NaNO3 was used as a control nitrogen source unrelated to purine transport activities in all tests scoring resistance/sensitivity to 5FU (rows 1–7). Rows 3–7 represent in vivo competition assays scoring the ability of excess purines (2 mM) to compete with the uptake of 5FU (100 μΜ), and thus revert 5FU sensitivity. Χ is xanthine, A is adenine, T is Thymine, HX is hypoxanthine, and U is uracil. Notice that T, HX, and U competed with 5FU uptake and suppressed sensitivity. Growth was also scored on minimal media containing UA (uric acid), X, or HX as sole nitrogen sources, none of which supported growth of the strains tested (three last rows). (C) Growth tests of A. nidulans of the same strains as in (A), on minimal media plus proline as a sole nitrogen source, supplemented or not with 100 mM NaCl. Notice that rSNBT1-N390T-mediated 5FU sensitivity is dependent on the presence of Na+ supplementation. Notice also that in the presence of proline as a nitrogen source, the wild-type rSNBT1 allele confers very moderate sensitivity to 5FU. (D) Left panel: 3H-uracil (0.1 μΜ) accumulation in strains expressing rSNBT1 and rSNBT1-N390T, performed in the presence or absence of 100 mM Na+, and in the presence or absence of excess (2 mM) unlabeled nucleobases, after a period of 10 min incubation with radiolabeled substrate. 3H-uracil accumulation in rSNBT1-N390T in the presence of 100 mM Na+ and absence of unlabeled nucleobase is arbitrarily taken as 100%. Right panel: relative 3H-uracil (0.1 μΜ) transport accumulation in Δ7 (negative control), rSNBT1, or rSNBT1-N390T strains as a time course. Uptake results are averages of three measurements for each concentration point. SD was 20%. (E) Inverted fluorescence microscopy images showing the subcellular localization of the GFP-tagged rSNBT1 and rSNBT1-N390T constructs. Notice that the strains used for microscopy are identical to those used in growth tests and uptake assays, as in all cases rSNBT1 sequences were tagged C-terminally with GFP (see Materials and Methods).