Figure 3.
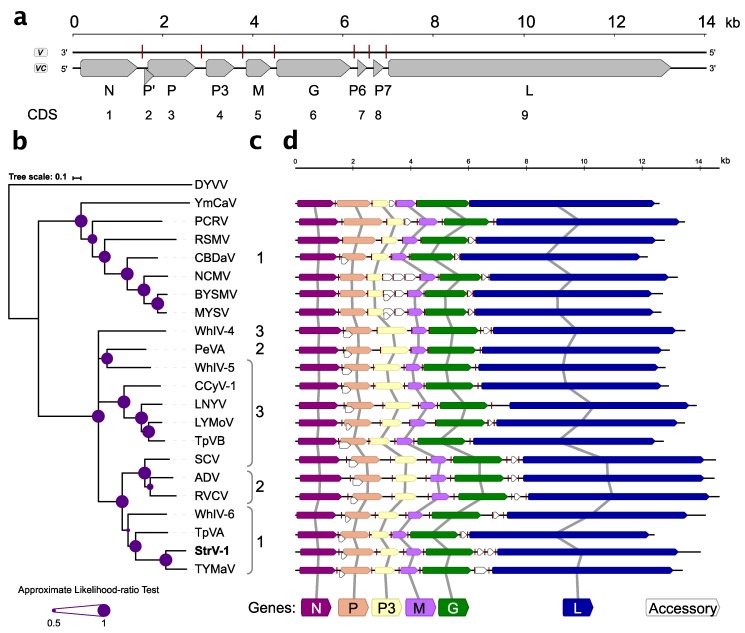
(a) Genomic organization of StrV-1-ČRM1, genotype A (MK211270). The nucleotide (nt) sequence is shown as a black line. Viral genomes and complementary strands are labelled as v and vc, respectively. Open reading frames (ORFs, or CDSs) are drawn as grey arrows, and putative regulatory regions are shown as red vertical lines. Segments are drawn to scale; (b) Phylogenetic tree based on L protein sequences of StrV-1 ČRM1 (B) and related cytorhabdoviruses. The acronyms are used as follows: alfalfa dwarf virus, ADV (NC_028237); barley yellow striate mosaic cytorhabdovirus, BYSMV (NC_028244); cabbage cytorhabdovirus 1, CCyV-1 (KY810772); colocasia bobone disease-associated virus, CBDaV (NC_034551); datura yellow vein nucleorhabdovirus, DYVV (NC_028231); lettuce necrotic yellows virus, LNYV (NC_007642); lettuce yellow mottle virus, LYMoV (NC_011532); maize yellow striate virus, MYSV (KY884303); northern cereal mosaic cytorhabdovirus, NCMV (NC_002251); papaya cytorhabdovirus, PCRV (KY366322); persimmon virus A, PeVA (NC_018381); raspberry vein chlorosis virus, RVCV (MK257717); rice stripe mosaic virus, RSMV (NC_040786); strawberry crinkle cytorhabdovirus, SCV (MH129615); tomato yellow mottle-associated virus, TYMaV (NC_034240); Trifolium pratense virus A, TpVA (MH982250); Trifolium pratense virus B, TpVB (MH982249); Wuhan Insect virus 4, WhIV-4 (NC_031225); Wuhan Insect virus 5, WhIV-5 (NC_031227); Wuhan Insect virus 6, WhIV-6 (NC_031232); yerba mate chlorosis-associated virus, YmCaV (KY366322). A plant-infecting nucleorhabdovirus, DYVV, was used as an outgroup. Scale bars refer to a phylogenetic distance expressed in amino acid substitutions per site. Support values are expressed graphically; (c) The number of predicted transmembrane domains (TM) in P’ proteins (when applicable); (d) Comparative genomic organization of StrV-1-ČRM1 (A) and other cytorhabdoviruses. Virus names are those from the phylogenetic tree. Shared gene synteny is shown with grey lines.