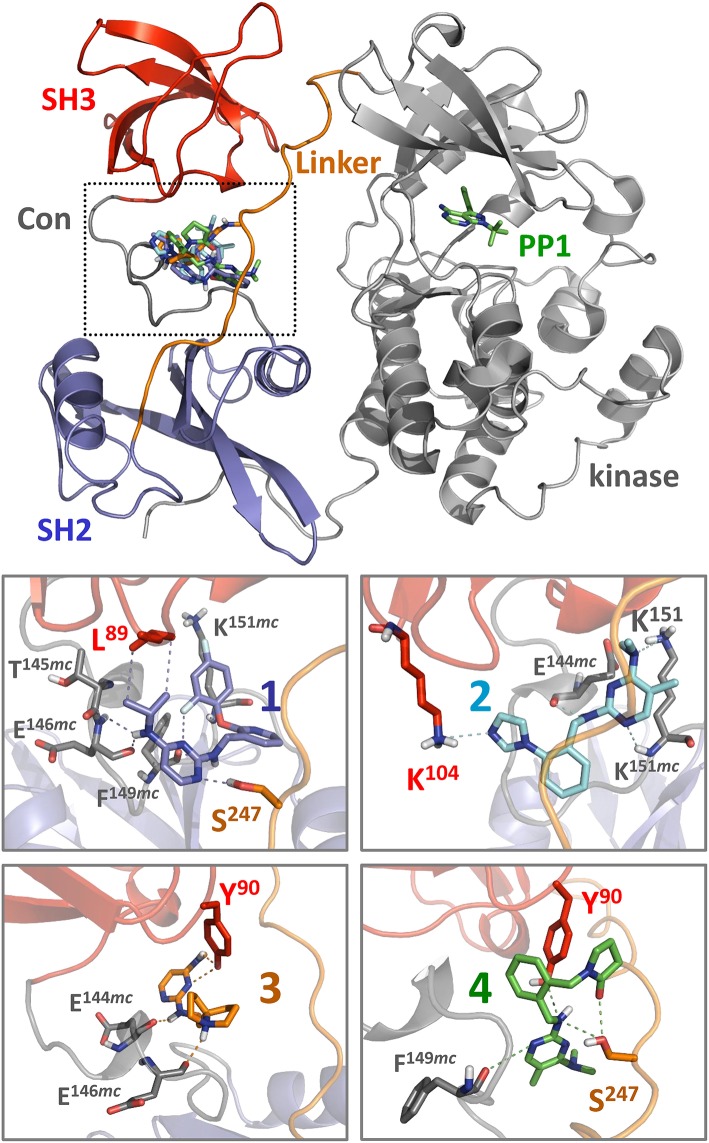
Figure 6.
Docking predicts binding of hit compounds to the Hck regulatory region. Compounds 1 through 4 from the Hck-U32L screen (Figure 4) were docked to the crystal structure of near-full-length Hck (PDB: 1QCF) using Smina (Koes et al., 2013). The overall near-full-length Hck structure is shown at the top including the kinase domain, where the ATP-competitive inhibitor PP1 marks the active site. All four compounds are predicted to bind within a pocket formed by the convergence of the SH3 domain, the SH3-SH2 connector (Con), and the SH2-kinase linker (boxed region). Close-up views of binding sites for compounds 1 through 4 (lower panels) show predicted interatomic contacts between the ligand and side/main chain residues ≤ 4 Å (dotted lines). Side chains and main chains (mc) predicted to participate in ligand binding are labeled. Nitrogen atoms present in the pyrimidine diamine core found in all four compounds make multiple contacts with the target protein. Compound 2 is unique among the four compounds, in that it makes predicted polar contacts with the side chain of SH3 Lys104 and the side and main chains of linker Lys151. This observation may relate to its greater stimulatory activity in the kinase activity assay compared to the other three compounds (Figure 7).