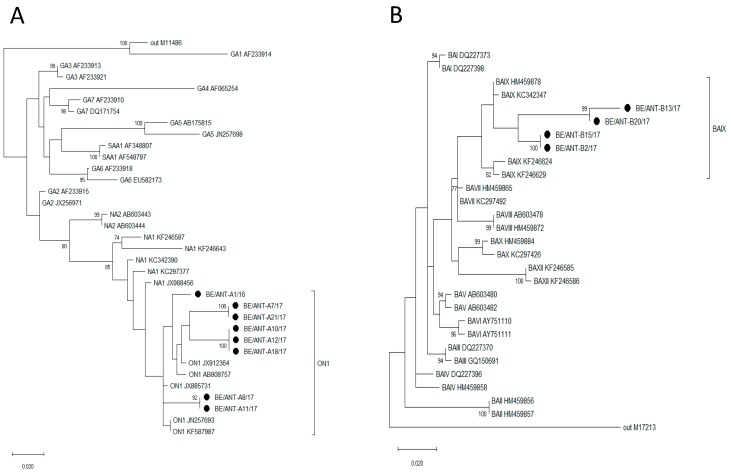
Figure 1.
Phylogenetic trees for RSV-A and RSV-B clinical isolates. The phylogenetic trees were constructed with maximum-likelihood with 1000 bootstrap replicates using MEGA X software. The trees are based on a 342nt and 330nt fragment of the G protein of RSV-A (A) and RSV-B (B) strains respectively, consisting of the second hypervariable region. Nucleotide sequences of the clinical isolates (indicated with •) were compared to reference strains found on GenBank (indicated with genotype and accession number). The outgroups are represented by prototype strains M11486 for RSV-A and M17213 for RSV-B. Bootstrap values greater than 70% are indicated at the branch nodes and the scale bar represents the number of substitutions per site.