Abstract
Heterochromatin protein 1 (HP1) facilitates the formation of repressive heterochromatin domains by recruiting histone lysine methyltransferase enzymes to chromatin, resulting in increased levels of histone H3K9me3. To identify chemical inhibitors of the HP1-heterochromatin gene repression pathway, we combined a cell-based assay that utilized chemical-mediated recruitment of HP1 to an endogenous active gene with high-throughput flow cytometry. Here we characterized small molecule inhibitors that block HP1-mediated heterochromatin formation. Our lead compounds demonstrated dose-dependent inhibition of HP1-stimulated gene repression and were validated in an orthogonal cell-based system. One lead inhibitor was improved by a change in stereochemistry, resulting in compound 2, which was further used to decouple the inverse relationship between H3K9 and H3K4 methylation states. We identified molecular components that bound compound 2, either directly or indirectly, by chemical affinity purification with a biotin-tagged derivative, followed by quantitative proteomic techniques. In summary, our pathway-based chemical screening approach resulted in the discovery of new inhibitors of HP1-mediated heterochromatin formation while identifying exciting new molecular interactions in the pathway to explore in the future. This modular platform can be expanded to test a wide range of chromatin modification pathways yielding inhibitors that are cell permeable and function in a physiologically relevant setting.
Keywords: epigenetics, high-throughput screening, small molecule, HP1, H3K9me3, gene repression
Introduction
Histone lysine methylation represents a critical posttranslational modification that regulates gene expression and is essential for proper tissue specialization during mammalian development.1,2 Disruptions to the careful balance of epigenetic pathways, such as histone lysine methylation, have recently been identified as drivers of human cancer.3–5 Histone lysine methylation can correlate with the activation or repression of gene transcription depending on the specific histone residue modified and the landscape of the chromatin where the histone lysine methylation is placed. For example, histone H3 lysine 4 trimethylation (H3K4me3) is typically found in promoters and gene bodies of active euchromatin genes, while H3K9me3 correlates with repressive heterochromatin.
Mammalian heterochromatin protein 1 (HP1) has three isoforms, HP1α, HP1β, and HP1γ, which contribute to the diverse functions of HP1. While these isoforms have been shown to mediate heterochromatin formation and facilitate gene repression, HP1 has been demonstrated to play key roles in transcription elongation and mRNA splicing.6–12 The chromodomain (CD) of HP1 allows for binding of H3K9me3, while the chromoshadow (CS) domain recruits in the histone methyltransferase enzymes Suv39H1/2, SETDB1, and G9a.13–15 These enzymes deposit H3K9me2/3 marks on neighboring histones as well as nonhistone substrates including p53, CDYL1, WIZ, ACINUS, and G9a.16–18 Subsequent binding of additional HP1 proteins facilitates the spreading of H3K9me3-marked heterochromatin, resulting in DNA compaction and gene repression.11 Thus, HP1 can function as a histone lysine methyl-reader and scaffolding protein facilitating the propagation of H3K9me3 to neighboring nucleosomes, which further spreads the heterochromatin domain and leads to gene silencing. Recently, heterochromatin domain formation was demonstrated to be mediated by the HUSH complex composed of TASOR, MPP8, and periphilin. This complex interacts with SETDB1 to deposit H3K9me3.19 The heterochromatin pathway is perturbed in a diverse set of human cancers, which makes it an exciting new pathway to consider targeting for future therapeutics.20–22
We developed the chromatin in vivo assay at the Oct4 locus (CiA:Oct4) to function as a modular recruitment platform for the study of epigenetic-modifying enzyme activity.23 This system takes advantage of the haplosufficient nature of the pluripotency factor OCT4 and replaces a single Oct4 allele with an enhanced green fluorescent protein (GFP) gene allowing for gene expression to be measured in single cells using flow cytometry. Upstream from the GFP protein are tandem arrays of zinc finger binding sites (ZFHD1) and Gal4 DNA binding domains. These domains served as the platform for recruiting epigenetic activities to the CiA:Oct4 locus using chemically induced proximity (CIP).24 A two-part recruitment tool using a Gal4-FK506 binding protein (Gal4-FKBP) and FKBP-rapamycin binding (FRB) fused to the CS domain of HP1α (FRB-csHP1α) was also stably transduced into the cell line. Upon CIP-rapamycin addition, Gal4-FKBP and FRB-csHP1α tethered csHP1α to the CiA:Oct4 locus, allowing for robust temporal control and visualization of heterochromatin formation dynamics. HP1 recruitment results in decreased GFP signal, removal of active H3K4me3, and deposition of H3K9me3, mirroring physiologic heterochromatin formation.23
In this study, we used the CiA:Oct4 system as a biological assay to identify pathway-based inhibitors of HP1-mediated heterochromatin. We conducted a novel small-molecule screen using a high-throughput flow cytometry-based approach as the primary readout. We identified several known and novel compounds that effectively inhibited the HP1-heterochromatin pathway, including UNC617 and UNC2524. Compounds were confirmed to decrease H3K9me3 levels, which was mechanistically consistent with inhibition of this pathway. Interestingly, we also observed a decoupling of the previously linked H3K9 and H3K4 methylation states upon compound treatment. We further determined through structure–activity relationship (SAR) studies that derivatives of UNC2524 were amenable to affinity purification followed by quantitative proteomic analysis using isobaric tags for relative and absolute quantitation (iTRAQ) labeling. This quantitative approach identified previously unexplored putative components of the HP1-heterochromatin pathway, including hepatoma-derived growth factor-related protein 2 (HDGFRP2). We present a modular approach that uses cell-based high- throughput screening to identify inhibitors that block an epigenetically active pathway acting on a native chromatin substrate in a physiologically relevant context.
Materials and Methods
Cell Culturing and Cell Lines
Mouse embryonic stem (ES) cells were adapted to grow on gelatin-coated plates without feeder cells in DMEM supplemented with 4.5 g/L glucose, 15% fetal bovine serum, L-glutamate, sodium pyruvate, HEPES buffer, NEAA, 2-mercaptoethanol, leukemia inhibitory factor (LIF), and penicillin/streptomycin (ES media) at 37 °C supplemented with 5% CO2. Second-generation lentiviral packaging vectors psPAX2, pMD2.G, and plasmid DNA were used to prepare virus as previously described.25 The CiA:Oct4 N118/N163 cell line containing viral integrations of N118 and N163 plasmids (N118, nLV EF-1α-Gal-FKBPx1-HA-PGK-Blast; N163, nLVEF-1α-HP1α(CS)-Frbx2(Frb+FrbWobb)-V5-PGK-Puro) was used for all experiments unless otherwise stated.23 CiA:Oct4 N118 was transduced with the lentiviral construct N192 (N192-nLV Dual Promoter EF-1α-MCS-PGK-Puro HP1γ (CS)-Frbx2(wobbmo)-V5) to yield the csHP1γ recruitment system CiA:Oct4 N118/N192. For the orthogonal recruitment system, a stable mouse ES cell line with a blue fluorescent protein (BFP) reporter gene with tetracycline response elements (TREs) was generated by recombinase-mediated cassette exchange, by introducing the reporter cassette DNA in plasmid YR06 into a gene trap located on chromosome 15 at genome coordinates chr15:99941948.26,27 Into this cell line, TetR-HP1-mCherry was introduced by lentiviral infection using nLV construct KS35(pEF1-TetR-HP1-P2A-mCherry).
Small-Molecule High-Throughput Screen and Flow Cytometry
On day 0, CiA:Oct4 N118/163 cells were grown in ES media and seeded at a density of 10,000 cells per well (100,000 cells/mL) into gelatin-coated 96-well plates. On day 1, media was aspirated and replaced with 100 μL of fresh ES media containing ±6 nM rapamycin and 10 μM dilution of compounds from the EpiG compound set. Media was exchanged with the same compound on day 2. On day 3, cells were washed with phosphate-buffered saline (PBS) and trypsinized using 0.25% trypsin-EDTA. Trypsin was quenched with serum. Cells were resuspended by pipetting to prepare the plates for flow cytometry analysis. A similar technique used for dose–response assays, SAR assays, and csHP1γ inhibition assay (only recruiting csHP1γ N192). Flow cytometry was performed using an Intellicyt iQue Screener (Albuquerque, NM) and analyzed with FlowJo software (Ashland, OR). Cell populations were gated as described in Supplemental Materials.
Orthogonal TetR-HP1 Recruitment Assay
TetR-HP1 cell lines were continuously grown with 1 μg/mL doxycycline to maintain an open chromatin state. On day 0, TetR-HP1 cells were grown in ES media containing doxycycline and seeded at a density of 10,000 cells per well in 96-well plates. On day 1, cells were washed with PBS and 100 μL of fresh ES media was added containing ± 1 μg/mL doxycycline and 5 μM compounds; this media was freshly added on day 2. On day 3, cells were trypsinized using 0.25% trypsin-EDTA. Trypsin was quenched with serum and then cells were analyzed by flow cytometry. The median BFP intensity was determined by FlowJo software analysis. Three biological replicates were averaged and used to generate standard error bars.
Compound 2 Activity and Fluorescence Microscopy
CiA:Oct4 N118/N163 cells were treated with ±7.5 μM compound 2 ± 6 nM rapamycin for 48 h. Cells were imaged in PBS using an Olympus IX71 microscope and analyzed using Cellsens software. Representative images were taken in two random plate locations for each of the four conditions. Image levels were normalized using Adobe Photoshop (San Jose, CA).
ChIP Sample Preparation and qRT-PCR
Chromatin immunoprecipitation (ChIP) was conducted essentially as previously described with the following antibodies: Abcam H3K9me3 (ab8898), Millipore HP1γ (05–690), Millipore H3K4me3 (05–745R), and Abcam EHMT2/G9a (ab40542).25 Samples were analyzed using the comparative △△Ct method and normalized against an intergenic control region or normalized to a housekeeping gene, intracisternal A-type particles (IAPs) (Suppl. Table S2).23,28 Experiments were performed in a minimum biological triplicate and data are representative of sample averages. t tests were used to determine significant p values.
Affinity Purification Sample Preparation and iTRAQ LC-MSIMS Analysis
Briefly, CiA:Oct4 N118/N163 nuclei were resuspended in 2 mL of shearing buffer (0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA [pH 8.0], 10 mM Tris HCl [pH 8.0]) and sonicated using a probe sonicator. Lysates were run over magnetic streptavidin beads (Dynabeads M-280 Streptavidin, Invitrogen, Carlsbad, CA) and washed and eluted as indicated. Samples were run on a 4%–20% tris-glycine gradient protein gel and then prepared for analysis. See Supplemental Materials for full proteomic experimental details.
Affinity Purification to Validate HDGFRP2 and MPP8 Binding
Hdgfrp2 (isoform b) was amplified from Mus musculus TC1 cDNA cloned into a pcDNA3.1-Myc-His expression vector at the NotI site by In-Fusion Cloning (Clontech, Mountain View, CA). Insert was then subcloned into lentiviral transfer plasmid nLV_N103 at the NotI site using In-Fusion. The final Hdgfrp2-Myc-His plasmid (S002) was transfected into 70%–80% confluent human embryonic kidney (HEK) Lenti-X 293T using PEI (1 μg/μL) and 25 μg of DNA. At 48 h, cells were trypsinized and pellets collected. Cells were lysed using M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA) supplemented with Benzonase (Sigma, St. Louis, MO) and pro tease inhibitors according to each manufacturer’s procedures. Cell debris was pelleted and supernatant lysates were transferred to microfuge tubes. Compounds 3 and 4 were incubated with magnetic streptavidin beads (Dynabeads M-280 Streptavidin) and washed in PBST (PBS 0.1% Tween-20). The beads-alone control was incubated with 3 mM D-biotin and washed in PBST. Beads coated with compounds 3 and 4 and the beads-alone control were incubated overnight at 4 °C with 1 mg of cell lysate. Beads were isolated using a magnet. Proteins bound to the magnetic beads were washed by vortexing with 50 mM HEPES, 150 or 500 mM NaCl, and 1% NP-40 three times. Sample supernatants were resuspended in 30 μL of 1× Laemmli loading dye and boiled for 5 min. A portion of each sample was run on a 4%–20% tris-glycine gradient protein gel and transferred for Western blot analysis to determine HDGFRP2 and MPP8 binding.
Histone Acid Extraction
Confluent CiA:Oct4 N118/N163 treated as indicated was trypsinized with 0.25% trypsin-EDTA. Nuclei were isolated by resuspending the pellet in lysis buffer (50 mM HEPES [pH 8.0], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton ×100) and incubating on ice for 10 min. Nuclei were pelleted at 6500g for 10 min at 4 °C and the supernatant was removed. Pellet was resuspended and rinsed in a second buffer containing 10 mM Tris (pH 8.0), 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA and centrifuged at 6500g for 5 min at 4 °C. The supernatant was removed, and the histones were extracted from the nuclei in 100–500 μL of 0.2 N HCl.29 Samples remained at 4 °C overnight and then were centrifuged at 6500g for 10 min at 4 °C. Supernatant containing histones was removed and neutralized with a 1/10 volume of 2 N NaOH, and protein concentration was determined by standard Coomassie Bradford assay (Pierce, Rockford, IL).
SDS-PAGE and Western Blot Analysis
Samples were prepared with 1× to 4× Laemmli loading buffer and boiled for 5 min before being loaded on 4%–20% tris-glycine gradient gels and transferred to a Millipore (Burlington, MA) Immobilon-FL polyvinylidene fluoride (PVDF) membrane. PVDF was blocked with Licor Odyssey Blocking Buffer (PBS) for at least 1 h. Primary antibodies (Active Motif anti-H3K9me3 39161, Active Motif anti-H3K9me2 39239, Active Motif anti-H4 61521, Proteintech anti-Mphosph8 16796–1-AP, EMD Millipore anti-Myc 05–724) were incubated overnight at 4 °C. Licor IRDye 680RD goat anti-mouse or Licor IRDye 800CW goat anti-rabbit secondary antibodies (1:15,000) were incubated for 30 min at room temperature. Western blots were imaged using the Licor Odyssey scanner and data were analyzed using Image Studio v5.2.
Chemical Synthesis for UNC2524 and Derivatives for SAR and Affinity Purification Studies
See Supplemental Materials for a detailed chemical synthesis and analytical data.
Results
Small-Molecule Screen for Epigenetic Modulators of the HP1-Heterochromatin Pathway
To identify modulators of the HP1-heterochromatin pathway, we employed a small-molecule screening approach, which used an epigenetic-targeted compound library (EpiG library) curated at the Center for Integrative Chemical Biology and Drug Discovery at the University of North Carolina (UNC). This set includes ~960 small molecules designed and synthesized to target diverse epigenetic regulatory proteins, making it an ideal small-molecule library to interrogate the HP1-heterochromatin pathway. While some compounds within the set have known biological activity, the majority do not. We developed a high-throughput flow cytometry-based screening platform that used the CiA:Oct4 system in a mouse ES cell line to express enhanced nuclear GFP as a reporter for chromatin dynamics.23 This approach allowed us to screen compounds in a high-throughput format and determine specific effects on the chromatin state with single-cell resolution in a temporally controlled manner.
In this study, we chemically induced HP1α recruitment to the Oct4 promoter region using the CiA:Oct4 system and measured gene activity by GFP expression. Lentiviral infection into the CiA:Oct4 cell line yielded stable expression of the Gal4-FKBP (N118) and FRB-csHP1α (N163) fusion proteins.23 Rapamycin addition tethers the FKBP and FRB domains, rapidly recruiting csHP1α to the CiA:Oct4 locus (Fig. 1a). HP1 recruitment was followed by removal of active chromatin marks such as H3K4me3, deposition of repressive H3K9me3, and gene repression. This method mimics the physiologic chromatin transformation that occurs at the Oct4 locus upon cellular differentiation of ES cells.
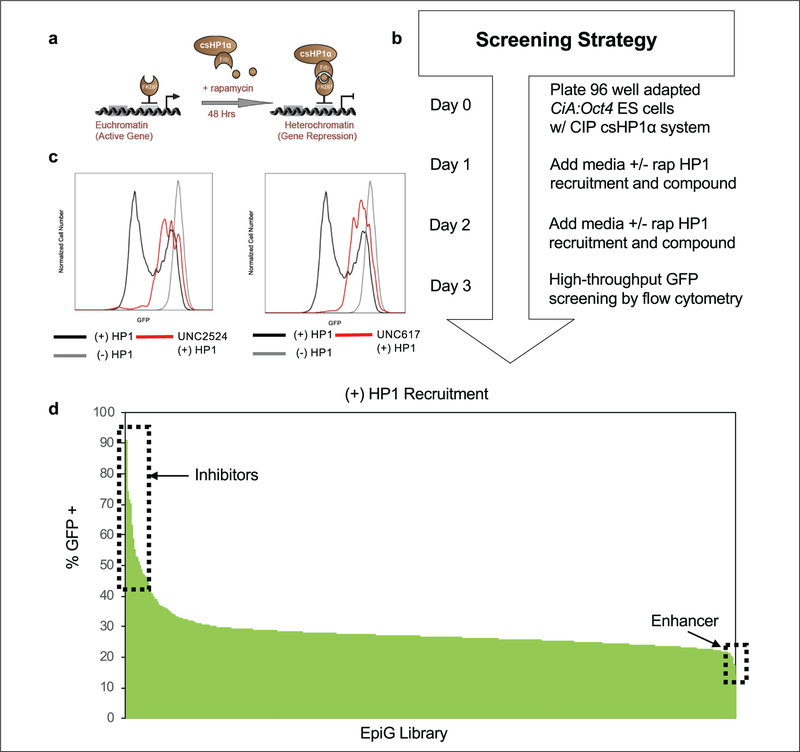
Figure 1.
High-throughput flow cytometry screen for modulators of HP1-mediated heterochromatin formation. (a) Cartoon of the CiA:Oct4 system utilizing CIP to recruit csHP1α. Addition of rapamycin facilitated the bridging of Gal4-FKBP and FRB-csHP1α fusions by CIP, resulting in HP1-heterochromatin formation and gene repression. (b) Diagram outlining the primary screening strategy workflow over a 3-day experimental time course. (c) Representative histogram of GFP fluorescence intensity for top inhibitors of HP1-mediated repression (UNC2524 and UNC617) at 10 μM compared with ±6 nM rapamycin controls. (d) Results of small-molecule screen showing the percentage of GFP-positive populations after 48 h of CIP-mediated HPI recruitment. Inset boxes highlight inhibitors or enhancers of HP1-mediated gene repression.
We screened the EpiG compound set at 10 μM with rapamycin and compound added at the same time 48 h prior to analysis by high-throughput flow cytometry using the CiA:Oct4 csHP1α recruitment cell line (Fig. 1b). The percentage of GFP-positive cells was determined by gating on the GFP-negative populations in control samples treated with rapamycin and dividing by total singlet cell counts (Suppl. Fig. S1a). In the primary screen to recruit csHP1α, 78 compounds had fewer than 200 cell events detected by flow cytometry and were removed due to lack of statistical confidence in the data. This lack of cells was likely due to compound toxicity at the 10 μM screening dose. Representative histograms of flow cytometry data showed that csHP1α recruitment facilitated by rapamycin addition (black line) resulted in cell populations shifting to be GFP negative. Cells that were not treated with CIP-rapamycin (gray line) remained near 100% GFP positive, as no csHP1α was recruited to the locus in these samples. Top inhibitors of the HP1-heterochromatin pathway, UNC2524 and UNC617 (red line), resulted in an increased expression of GFP despite csHP1α recruitment, which led to a greater percentage of GFP-positive cells (Fig. 1c). Remaining compounds were arranged from highest to lowest percentage GFP-positive population (Fig. 1d). We identified 34 compounds that were 2 standard deviations above the mean, with UNC617 and UNC2524 representing the two compounds that resulted in the largest increase in GFP expression. Compound UNC00000202 demonstrated an ability to enhance HP1 pathway repression with nearly 10% less GFP expression than controls. However, we chose to pursue the inhibitors of HP1-mediated gene repression in this study. Enhancers of the HP1-heterochromatin pathway represent candidates for potential future study.
The CiA:Oct4 csHP1α recruitment system also functioned as an internal counterscreen for compounds that cause a decrease in GFP expression, likely due to toxicity or cell differentiation, independent of rapamycin-recruited csHP1α. ES cell differentiation causes the Oct4 locus to be silenced, resulting in a decrease in GFP expression. We screened the EpiG compound set at 10 μM without rapamycin-induced csHP1α recruitment for 48 h prior to analysis by high-throughput flow cytometry. Lack of csHP1α recruitment resulted in nearly 100% GFP-positive cells in controls and with most compounds (Suppl. Fig. S1b). From the EpiG library, 72 compounds were identified below the 200-event cutoff in the minus rapamycin counterscreen and could not be interpreted. Compounds that resulted in a greater than 10% reduction in GFP-positive cell populations compared with the mean were removed from the data set as Oct4 repression is a sign of cellular differentiation. This cutoff allowed us to conservatively eliminate compounds that are causing Oct4 repression independent of HP1 recruitment, while keeping the most active compounds for further downstream analysis (Suppl. Fig. S1c). Lead inhibitors of HP1-mediated heterochromatin formation, UNC2524 and UNC617, did not decrease GFP-positive population levels, but demonstrated increased expression of GFP compared with controls, indicating greater gene activation (Suppl. Fig. S1d). Only compounds that did not cause CiA:Oct4 repression independent of csHP1α recruitment or result in cell toxicity in either the plus or minus rapamycin screens were used to generate the final data set for the screen (Suppl. Fig. S1e). Supplemental Table S1 summarizes the key parameters of this small-molecule screen.
Functional Analysis of Lead Screen Compounds
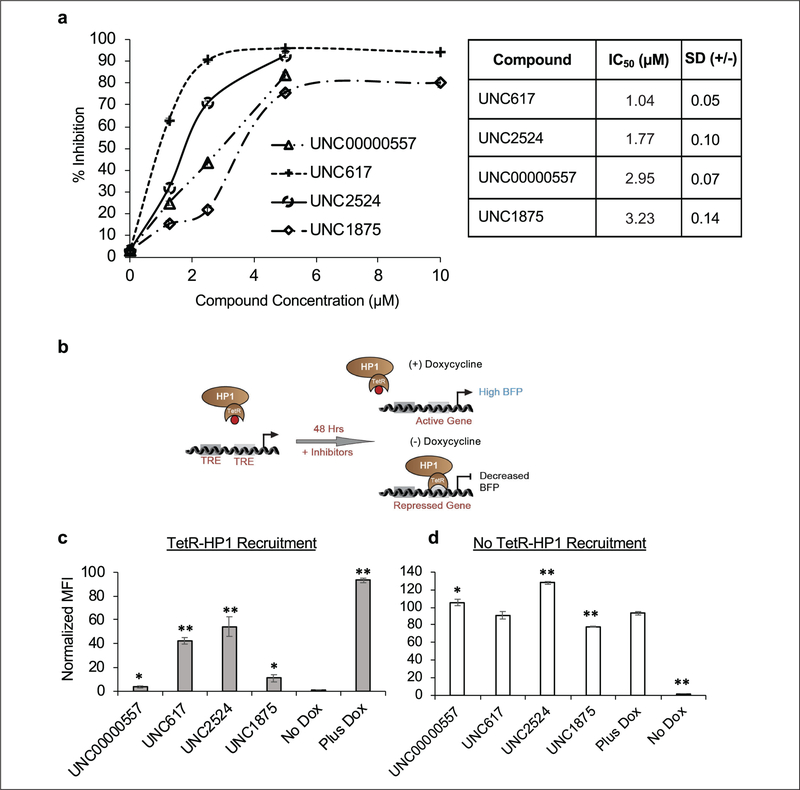
To confirm the activity of our lead compounds, we first performed a dose–response curve at 10, 5, 2.5, 1.25, and 0 μM of each compound with and without rapamycin to determine the maximum biological activity of the compounds without compromising cell viability. Like our screening approach, CiA:Oct4 csHP1α-recruiting cells were cultured with and without rapamycin-mediated HP1 recruitment. The response curves demonstrated dose-dependent activity in our biological assay, correlating increased compound concentration with increased inhibition of the HP1 pathway. UNC617, previously reported as a modest inhibitor of G9a/GLP repression in a cell-based screen,30 demonstrated the most potent response in our assay (IC50 1.04 μM), followed by UNC2524, UNC00000557, and UNC1875 with IC50 values of 1.77, 2.95, and 3.23 μM, respectively (Fig. 2a). Expanded dose-response curves for 12 of the top inhibitors were also conducted with HP1 recruitment (Suppl. Fig. 2a) and without HP1 recruitment (Suppl. Fig. 2b). Absent data points were caused by compound toxicity, resulting in too few cells to interpret the data with confidence. We concluded from these data that a 5–10 μM dose maximized compound assay activity and cell viability for our top hits from the screen.
Figure 2.
Hit compounds demonstrate dose-dependent response and are validated by an orthogonal TetR-HP1 recruitment system. (a) Dose-response curves were conducted for the top four hit compounds (UNC00000557, UNC617, UNC2524, and UNC1875). CiA:Oct4 N118/N163 cells were treated with compound at 10, 5, 2.5, 1.25, and 0 μM doses over 48 h ± 6 nM rapamycin. Flow cytometry analysis was used to determine the percentage of GFP-positive cells, and those values were converted to percent inhibition. IC50 values are displayed in the associated table. (b) Cartoon depicting the TetR-HP1 orthogonal recruitment system and the outcome of BFP expression after 48 h of inhibitor treatment ± 1 μg/mL doxycycline. Normalized levels of median BFP expression after 48 h of 5 μM compound treatment with (c) and without (d) HP1 recruitment. t test statistical analysis in panel c compared samples to the No Dox control, while in panel d it compared samples to the Plus Dox control. n ≥ 3; *p ≤ 0.05, **p ≤ 0.0I.
To exclude the possibility that our lead compounds were inhibiting CIP-rapamycin recruitment of csHP1α to the CiA:Oct4 locus, or that HP1-heterochromatin inhibition was specific to Oct4, we sought to confirm the function of our lead compounds in an orthogonal recruitment system located at a distinct genetic locus devoid of epigenetic marks. HP1 was fused to the tetracycline repressor (TetR) and stably expressed in a mouse ES cell line expressing a BFP reporter gene upstream of the TRE. Like rapamycin addition, the absence of doxycycline causes TetR-HP1 to be recruited to the TRE, resulting in HP1-heterochromatin formation. The addition of doxycycline inhibits TetR from binding to TRE, leading to gene activation (Fig. 2b). We tested compounds UNC00000557, UNC617, UNC2524, and UNC1875 at 5 μM for 48 h with and without TetR-HP1 recruitment. All compounds tested were shown to significantly inhibit HP 1-mediated heterochromatin formation, with UNC617 and UNC2524 remaining the most potent inhibitors (Fig. 2c). Further, only UNC1875 demonstrated a minimal decrease in median BFP expression independent of TetR-HP1 recruitment, while all other compounds resulted in either no change or increased BFP expression compared with controls, indicating an increase in gene expression (Fig. 2d). These data corroborated our primary screen results and demonstrated, using an orthogonal recruitment method, that the lead compounds did not inhibit tethering of HP1 by rapamycin. Further, lead compounds were capable of inhibiting HP1-mediated gene repression at a second gene locus measured by a different fluorescent reporter.
HP1-mediated gene repression and silencing is mediated by H3K9me3 deposition. We sought to determine if our lead compounds were not only affecting gene repression at specific genetic loci, but also affecting global H3K9me levels. csHP1α-recruiting CiA:Oct4 cells were treated with 10 μM compound for 48 h. After 48 h of treatment, histones were isolated from nuclei by acid extraction. H3K9me2 and H3K9me3 levels were assayed by Western blot and normalized to histone H4 as a loading control (Suppl. Fig. 3a). Quantification of H3K9me2 and H3K9me3 Western blots demonstrated that all lead compounds trended toward decreased global H3K9me2/3, though only UNC00000557 and UNC617 showed statistically significant reduction in H3K9me2 levels. UNC2524 and UNC1875 also decreased H3K9me2/3, but not to the significant extent of the other compounds (Suppl. Fig. 3b).
HP1 has three isoforms, HP1α, HP1β, and HP1γ. HP1α and HP1γ were previously identified to function similarly upon recruitment to the CiA:Oct4 locus, whereas HP1β was not able to appreciably repress the reporter allele (unpublished observations). We therefore wanted to determine percentage inhibition upon recruitment of csHP1γ for the best 18 compounds identified in the primary screen. We transduced CiA:Oct4 ES cells containing the Gal4-FKBP (N118) plasmid with the FRB-csHP1γ (N192) vector using lentivirus to yield a stable CiA:Oct4 CIP-controlled csHP1γ-recruiting cell line. After 48 h of treatment with 5 μM compound with and without csHP1γ recruitment, the percentage of GFP-positive cells was determined by flow cytometry analysis. We identified that like the csHP1α recruitment and inhibition screen, UNC617 and UNC2524 were the most potent inhibitors of csHP1γ enrichment at the CiA:Oct4 locus. Nearly all lead compounds demonstrated significant inhibition of csHP1γ-mediated gene repression at the CiA:Oct4 locus (Suppl. Fig. 4a). Additionally, very low compound toxicity and independent repression of the CiA:Oct4 allele was observed, with the greatest reductions being ~3% and 4% for UNC1868 and UNC1871 (Suppl. Fig. 4b). The reproducible inhibition of functionally similar HP1 isoforms contributes to the robustness of our assay and provides greater evidence that our top hits represented novel inhibitors of the HP1-heterochromatin pathway.
Structure–Activity Relationship of UNC2524
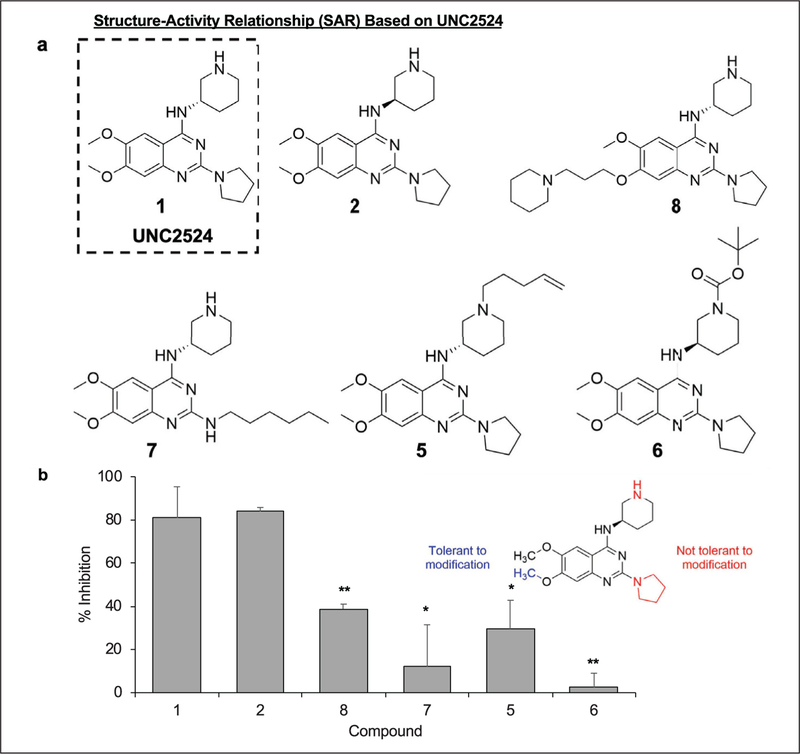
Despite elucidating the general phenotypes of our new HP1-heterochromatin pathway inhibitors with respect to chromatin state, we did not understand how our compounds were inhibiting HP1-mediated gene repression. UNC2524 was chosen for further investigation due to its potent inhibitory phenotype in our assays, in addition to having no known targets or function. SAR studies were performed with UNC2524 to accomplish two goals: first, to optimize compound activity in the CiA:Oct4 assay, and second, to determine if UNC2524 was tolerant to side-chain modification for downstream chemical affinity proteomic experimentation to identify the HP1-heterochromatin pathway molecular components that interact with UNC2524.
Compounds 2, 5, 6, 7, and 8 were chemically synthesized as close analogs of UNC2524 (compound 1) (Fig. 3a). Detailed chemical synthesis schemes can be found in the Materials and Methods section. To ascertain the biological activity of these chemical derivatives, we used the CiA:Oct4 csHP1α recruitment system. Cells were treated with 5 μM of each compound as indicated for 48 h prior to analysis by flow cytometry. Compound activity was confirmed by an increase in percent inhibition compared with untreated control samples. Compounds 5 and 6 added a pentene chain and Boc group to the piperidine amine, respectively, while compound 7 substituted a hexyl chain for the pyrrolidine. These modifications resulted in a decrease in inhibition for compounds 5 and 6 and an increase in cell toxicity for compound 7, indicating that these regions of the parent molecule, 1, were not optimal places to attach an affinity handle (Fig. 3b). We identified that the 7-methoxy side chain was most amenable to modification with a bulky group, as compound 8 contains an alkyl piperidine substituent at this position and preserves 50% biological activity of compound 1 with minimal cell toxicity. Finally, a small increase in activity over UNC2524 was observed with compound 2 by changing the stereochemistry of the piperidine ring. Considering these data, we used compound 2 as a scaffold for incorporating a biotin tag onto the 7-methoxy side chain of the compound.
Figure 3.
SAR study of compound 1 (UNC2524). (a) A series of analogs of compound 1 (UNC2524) were designed to optimize compound activity and determine the amenability of compounds for biotinylation for affinity purification methods. (b) CiA:Oct4 csHP1α-recruiting cells were treated with compounds at 5 μM for 48 h ± 6 nM rapamycin. Flow cytometry analysis was used to determine the percentage of the GFP-positive cell population. Compound 7 treatment resulted in toxicity and low cell counts. These data were normalized to percent inhibition compared with untreated controls. n ≥ 3; *p ≤ 0.05, **p ≤ 0.01.
Compound 2 Inhibits HP1-Mediated Gene Repression
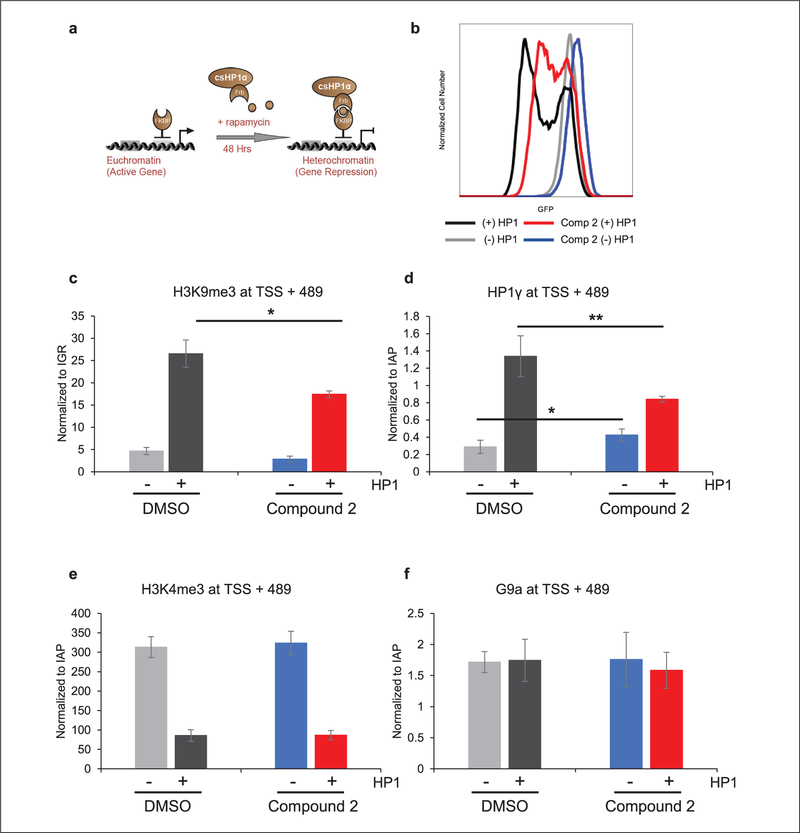
We endeavored to confirm the inhibition of H3K9me3 deposition by compound 2 to ensure that it was functioning similarly to UNC2524. CiA:Oct4 csHP1α recruitment cells were treated with compound 2 for 48 h with and without rapamycin mediating HP1 recruitment (Fig. 4a). Representative brightfield and fluorescence images were taken of the four sample types: (−) HP1/(−) compound 2, (−) HP1/( + ) compound 2, ( + ) HP1/(−) compound 2, and ( + ) HP1/( + ) compound 2. ES cell colony morphology was normal in all samples, though rapamycin did result in a slightly decreased colony size. When compared with controls, compound 2 did not result in gross cell differentiation or affect colony morphology in terms of colony shape and granularity observed by light microscopy (Suppl. Fig. S5). Subsequently, all samples were analyzed by flow cytometry to confirm the inhibitory effects of compound 2 on HP1-mediated gene repression. Figure 4b is a representative histogram that confirmed that compound 2 resulted in increased GFP expression with (red) and without (blue) HP1 recruitment compared with untreated controls.
Figure 4.
Compound 2 inhibits HP1-mediated gene repression. (a) Cartoon of the CiA:Oct4 system utilizing CIP to recruit csHP1α. The addition of rapamycin facilitated the bridging of the Gal4-FKBP and FRB-csHP1α fusion, resulting in HP1-heterochromatin formation and gene repression. (b) CiA:Oct4 csHP1α-recruiting cells were incubated with 7.5 compound 2 ± 6 nM rapamycin for 48 h. GFP expression was analyzed by flow cytometry. Following 48 h of treatment with compound 2 ± 6 nM rapamycin, ChIP was performed to determine enrichment levels of (c) H3K9me3, (d) HPI γ, (e) H3K4me3, and (f) G9a at +489 bp downstream of TSS. n ≥ 3; *p ≤ 0.05, **p ≤ 0.01.
ChIP was utilized to determine the levels of H3K9me3 at the CiA:Oct4 locus upon treatment with compound 2. We previously demonstrated that enrichment of H3K9me3 was greatest between 400 and 700 base pairs (bp) downstream of the transcriptional start site (TSS).23 We analyzed the levels of H3K9me3 at 489 bp downstream of the TSS in all four sample conditions. Real-time quantitative PCR (qRT-PCR) was used to amplify this region, and CT values were normalized to an intergenic region using the △△Ct method. Samples lacking HP1 recruitment, as expected, did not show enrichment in H3K9me3 due to lack of csHP1α recruitment. Conversely, H3K9me3 increased ~5.7-fold when treated with rapamycin, leading to csHP1α recruitment. Addition of compound 2 decreased the enrichment of H3K9me3 by ~34% compared with the plus rapamycin control samples (Fig. 4c).
To further characterize the effects of compound 2 on the HP1-heterochromatin pathway, we assayed for perturbations in the levels of HP1γ, H3K4me3, and G9a at the CiA:Oct4 locus using ChIP followed by qRT-PCR analysis. Enrichment levels were determined at 489 bp downstream of the TSS and normalized to a housekeeping gene, IAPs. As shown previously, HP1α recruitment leads to endogenous HP1γ binding to the CiA:Oct4 locus as a heterochromatin domain is formed. Because csHP1α is being actively recruited to the CiA:Oct4 locus, we chose to measure endogenous HP1γ levels to determine if HP1 recruitment was inhibited by compound 2. Despite a small increase in HP1γ caused by compound 2 alone, HP1γ levels were shown to significantly decrease (37%) upon HP1-mediated gene repression in the presence of compound 2 compared with control samples with HP1 recruitment alone (Fig. 4d). The histone mark H3K4me3 is associated with active gene transcription and was previously reported to decrease upon HP1-mediated gene repression.23 H3K4me3 levels were enriched ~3.7-fold in samples lacking HP1 recruitment, as expected. Surprisingly, no change in H3K4me3 levels was detected upon compound 2 treatment despite the decrease in H3K9me3 (Fig. 4e). Finally, the histone lysine methyl-transferase enzyme G9a was demonstrated to be required for silencing of Oct4 during cell differentiation and development.31 We assayed G9a levels to determine if G9a is contributing to HP1-mediated gene repression in our inhibition assay. No significant changes in G9a levels were observed across the four treatment conditions, indicating that G9a either is not recruited to the CiA:Oct4 locus under our assay conditions or is not detected by our assay (Fig. 4f). These data corroborated the microscopy and flow cytometry data that demonstrated an increase in GFP expression due to inhibition of endogenous HP1γ recruitment, which coincided with a decrease in the repressive H3K9me3 mark.
Components of HP1-Heterochromatin Pathway Identified by Chemical Proteomics
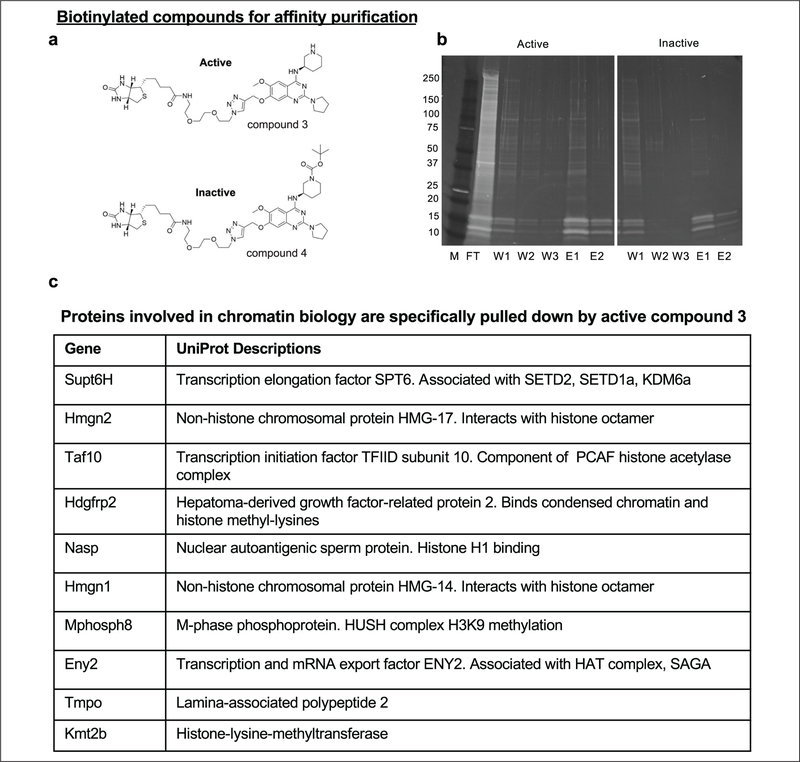
Compound 2 is a novel small molecule that we have demonstrated to inhibit the HP1-heterochromatin pathway for gene repression. To identify cellular targets of compound 2, we combined chemical affinity purification and quantitative mass spectrometry approaches. Biotin-tagged compound 2 (compound 3) was created by the addition of a biotin moiety off the 7-methoxy side chain of compound 2 and used as our active affinity reagent (Fig. 5a). Biotin-tagged compound 6 (compound 4) contained an inactivating Boc group to function as a negative control affinity reagent (Fig. 5a). Additional negative control samples included beads alone and preincubation of the nuclear lysates with excess compound 2 with the expectation that compound 2 would block pulldown of the cellular target(s) by compound 3.
Figure 5.
Chemical proteomics identifies putative HPI-heterochromatin pathway components. (a) Compound 3 (active) and compound 4 (inactive control) were used for chemical affinity purification to identify binding targets of compound 2 from CiA:Oct4 sonicated nuclear lysates. (b) Tris-glycine gradient protein gels (4%–20%) stained with Sypro Ruby stain loaded with marker (M), flow-through (FT), wash 1 (W1), wash 2 (W2), wash 3 (W3), elution 1 (E1), and elution 2 (E2) from affinity pulldown experiments comparing compounds 3 and 4. (c) The table lists select proteins enriched in the compound 3 sample (active) compared with controls as identified by iTRAQ LC-MS/MS with a summary of their UniProt descriptions.
Nuclei were harvested from CiA:Oct4 cells followed by lysis and shearing of genomic DNA by probe sonication to decrease sample viscosity and aid in protein dissociation from chromatin. Nuclear lysates were incubated with active (compound 3) and inactive (compound 4) biotin-tagged compounds. Magnetic streptavidin beads were used to isolate proteins bound to the biotinylated compounds. Samples were washed in buffer containing 150 mM NaCl, and bound proteins were eluted with excess compound 2 and finally with 3 mM D-biotin. Pulldown fractions from compounds 3 and 4 were run on 4%–20% tris-glycine gels and stained with Sypro Ruby stain for visualization (Fig. 5b).
Elution fractions were precipitated, and samples were prepared for quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using iTRAQ.32 Quantitative analysis identified proteins enriched in the active affinity purification sample compared with the various negative controls. Proteins that had greater than two unique peptides and a sample-to-control ratio of 1.4 or greater were considered for further analysis. Several proteins that bound to the active compound are known to play a role in chromatin biology. These include novel contributors to the HP1-heterochromatin pathway, such as SUPT6H, HMGN2, TAF10, HDGFRP2, NASP, HMGN1, ENY2, TMPO, and KMT2B, as well as MPHOSPH8 (MPP8), a known member of the HUSH complex, which contributes to H3K9 methylation and heterochromatin (Fig. 5c).19
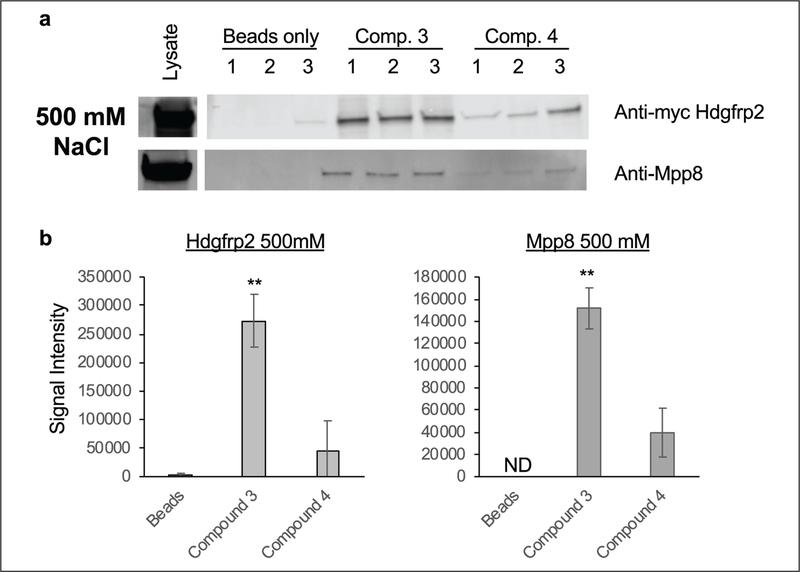
We sought to validate these findings by assaying the interaction of HDGFRP2 and MPP8 with compounds 3 (active) and 4 (inactive) under increasingly stringent wash conditions. HEK Lenti-X 293T cells were transfected with an HDGFRP2-Myc-His construct and allowed to express for 48 h prior to the harvesting of cell lysates. Streptavidin beads alone or conjugated to compounds 3 or 4 were incubated with cell lysate and subsequently washed with 150 or 500 mM NaCl-containing buffer prior to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. Washing conditions with 150 mM NaCl demonstrated a ~1.5-fold increase in copurification of HDGFRP2 to compound 3 and 4 compared with beads-alone controls. MPP8 showed ~3.1-fold increased binding to the active compound 3 compared with a ~1.9-fold increased affinity pulldown to the inactive compound 4 (Suppl. Fig. S6a). Upon the wash concentration being increased to 500 mM NaCl, significant enrichment of HDGFRP2 and MPP8 binding to the active compound 3 was observed. HDGFRP2 copurified with compound 3 ~100-fold more than the beads alone and ~6-fold higher than the inactive control, while MPP8 preferentially bound the active compound by ~3.8-fold compared with the inactive control (Fig. 6a). Enrichment over beads alone could not be determined for MPP8 because there were no visible protein bands present in the beads-alone samples. These data validate the iTRAQ proteomics for MPP8 and HDGFRP2 and suggest either direct binding to compound 3 or an indirect binding mechanism via yet to be elucidated protein complexes.
Figure 6.
Compound 3 chemiprecipitates HDGFRP2 and MPP8. HEK Lenti-X 293T cells were transfected with Myc-His-tagged Hdgfrp2 (S002) and expressed for 48 h prior to cell lysate being harvested. M-280 streptavidin beads alone or conjugated with compound 3 or 4 were incubated with 1 mg of lysate overnight. (a) Samples were washed three times with 500 mM NaCl-containing buffer. Samples were boiled in 1 × Laemmli buffer and run on 4%–20% tris-glycine gradient gels and transferred to PVDF to determine levels of HDGFRP2 and MPP8 by Western blot analysis using Licor Odyssey. (b) Band signal intensity is averaged and quantitated below. ND = not determined; n = 3; *p ≤ 0.05, **p ≤ 0.01.
Discussion
Disruption of epigenetic pathways has been demonstrated to drive diverse classes of human cancer. Components of the HP1-heterochromatin pathway have been identified as dysregulated in breast, uterine, prostate, and pancreatic carcinomas.20 Overexpression of pathway components is correlated with poor outcomes for patients with breast and liver cancer.20,33 Though inhibition of heterochromatin could be an attractive therapeutic strategy, currently no FDA-approved therapeutic targeting the HP1-heterochromatin pathway exists.
Here we present a novel pathway-based small-molecule high-throughput screening approach that identified modulators of HP1-mediated heterochromatin formation. Primary and secondary screens with and without rapamycin-induced HP1 recruitment allowed for us to internally control for and eliminate compounds that caused a decrease in GFP expressi on independent of HP1 recruitment. Top inhibitor compounds included UNC00000557, UNC617, UNC1875, and UNC2524. UNC617 was reported as an inhibitor of the histone lysine methyltransferase G9a, which functions by adding the H3K9me2 mark.30 Inhibition of H3K9me2 deposition would result in a decreased ability to add the H3K9me3 mark necessary for HP1-heterochromatin formation. Identifying relevant inhibitors like UNC617 validated our approach to identify modulators of the HP1-heterochromatin pathway. Each of our lead compounds possessed IC50 values of 1–4 μM, while UNC617 and UNC2524 were most potent. Top screen compounds were further shown to inhibit the csHP1γ isoform in a similar manner to csHP1α. UNC617 and UNC2524 remained the most effective inhibitors of the 18 top compounds screened against csHP1γ recruitment.
As a second counterscreen, we employed an orthogonal recruitment method using a TetR-HP1 fusion at a distinct genomic locus. As in our original screen, lead compounds demonstrated similar capacities to inhibit HP1-mediated gene repression. These data demonstrated that our compounds inhibit HP1-mediated gene repression independent of the recruitment method and at distinct genetic loci. To further expand the scope of activity for our compounds to the global cell level, we determined H3K9me2/3 levels on acid-extracted histones by Western blot. We determined that our top inhibitor compounds trended toward decreased whole-cell H3K9me2 and H3K9me3 levels, and that significant reductions were measured for a subset of compounds. We found that measuring the steady-state kinetics of histone marks after treatment was confounded due to balancing compound activity with compound toxicity. The trend toward a reduction in global methylation levels indicated that the compounds not only were functioning at the specific genetic loci tested but also are global inhibitors of the HP1-heterochromatin pathway for gene repression.
SAR studies identified compound 2 as our most potent inhibitor of HP1-mediated gene repression. Using ChIP analysis, we characterized the CiA:Oct4 allele to determine the effect of compound 2 treatment on HP1-heterochromatin pathway function. Compound 2 similarly inhibited H3K9me3 deposition and HP1γ localization. By inhibiting HP1 recruitment during HP1-mediated gene repression, H3K9me2/3 levels were expected to decrease due to a lack of recruitment scaffold for the histone lysine methyltransferase enzymes. H3K4me3 was previously shown to exist in an inverse relationship to H3K9me3. Surprisingly, we did not observe an increase in H3K4me3 as H3K9me3 decreased with compound 2 treatment. Compound 2 treatment allowed for the separation of two previously linked epigenetic marks and may represent a means to study this interaction in the future.
The phenotypic results of compound 2 treatment may not solely be a result of only direct inhibition of HP1 or a known component of the heterochromatin pathway, but an indirect effect leading to inhibition of gene repression. Recognizing this, we felt that employing a pathway-based screening approach combined with chemical affinity purification strategies and iTRAQ quantitative MS/MS would enable us to detect direct and indirect protein and protein complex interactions involved in HP1-heterochromatin gene repression. We identified SUPT6H, HMGN1, HMGN2, TAF10, NASP, ENY2, MPHOSPH8 (MPP8), KMT2B, TMPO, and HDGFRP2 as enriched compared with control samples. To validate the proteomic results, we assayed enrichment levels of one known and one unknown interacting protein, MPP8 and HDGFRP2, respectively, when pulled down by an active compound compared with an inactive compound or beads-alone controls under increasing salt concentrations. Under stringent 500 mM NaCl wash conditions, both MPP8 and HDGFRP2 were enriched in the active compound pulldown. Increased stringency may have eliminated competing binding partners or changed protein structures, allowing for greater engagement of the active compound compared with inactive control. It remains possible that MPP8 and HDGFRP2 act indirectly in tightly bound protein complexes, which facilitated the binding of compound 3 (biotinylated compound 2) and were indirectly pulled down. From these data, we suggest that HDGFRP2 could represent a component of the HP1-heterochromatin gene repression pathway. HDGFRP2 is closely related to the lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75), which is known to bind the integrase enzyme of HIV, leading to incorporation into active regions of chromatin.34 HDGFRP2 contributed to the efficiency and specificity of HIV integration, but prefers binding repressed chromatin marks.35 In addition to viral integration, HDGFRP2 was reported to interact with HP1β (CBX1) during DNA repair of silenced genes.34
In conclusion, we presented a modular high-throughput flow cytometry-based screening approach that resulted in the discovery of novel small-molecule inhibitors of HP1-mediated heterochromatin gene repression. Our chemical genetic approach identified new components to a critical pathway in mammalian development. These new tools will help to generate a better model of the molecular order of events for heterochromatin gene repression during development and disease. This unique approach can be easily adapted to identify inhibitors of other mammalian epigenetic pathways in the physiologic setting of a primary cell line.
Supplementary Material
Acknowledgments
We thank Drs. S. Frye, L. James, and W. Jansen from the Center for Integrative Chemical Biology and Drug Discovery (CICBDD) at The Eshelman School of Pharmacy for providing us with the EpiG compound plates and helpful discussion. We gratefully acknowledge L. James for critical reading of the manuscript.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: N.A.H. gratefully acknowledges support of this work from the University Cancer Research Fund, University of North Carolina at Chapel Hill; an American Association of Colleges of Pharmacy new investigator award; an Eshelman Institute for Innovation award; and grant R01GM118653 from the U.S. National Institutes of Health. J.J. gratefully acknowledges support from grants R01CA230854, R01GM122749, R01CA218600, and R01HD088626 from the U.S. National Institutes of Health. K.V.B. acknowledges support from an American Cancer Society postdoctoral fellowship PF-14–021-01-CDD. The CICBDD was supported by the National Institute of General Medical Sciences, National Institutes of Health grant R01GM100919, and the University Cancer Research Fund, University of North Carolina at Chapel Hill. The UNC Flow Cytometry Core Facility is supported in part by the P30 CA016086 Cancer Center Core support grant to the UNC Lineberger Comprehensive Cancer Center and in part by the North Carolina Biotech Center institutional support grant 2015-IDG-1001. This research is based in part on work conducted using the UNC Proteomics Core Facility, which is supported in part by the P30 CA016086 Cancer Center Core support grant to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplemental material is available online with this article.
References
- 1.Greer EL; Shi Y Histone Methylation: A Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black JC; Van Rechem C; Whetstine JR Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol. Cell 2012, 48, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrowsmith CH; Bountra C; Fish PV; et al. Epigenetic Protein Families: A New Frontier for Drug Discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [DOI] [PubMed] [Google Scholar]
- 4.Dawson MA; Kouzarides T Cancer Epigenetics: From Mechanism to Therapy. Cell 2012, 150, 12–27. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald IA; Hathaway NA Epigenetic Roots of Immunologic Disease and New Methods for Examining Chromatin Regulatory Pathways. Immunol. Cell Biol. 2015, 93, 261–270. [DOI] [PubMed] [Google Scholar]
- 6.Moazed D Common Themes in Mechanisms of Gene Silencing. Mol. Cell 2001, 8, 489–498. [DOI] [PubMed] [Google Scholar]
- 7.Saint-Andre V; Batsche E; Rachez C; et al. Histone H3 Lysine 9 Trimethylation and HP1γ Favor Inclusion of Alternative Exons. Nat. Struct. Mol. Biol. 2011, 18, 337–344. [DOI] [PubMed] [Google Scholar]
- 8.Yearim A; Gelfman S; Shayevitch R; et al. HP1 Is Involved in Regulating the Global Impact of DNA Methylation on Alternative Splicing. Cell Rep. 2015, 10, 1122–1134. [DOI] [PubMed] [Google Scholar]
- 9.Lev Maor G; Yearim A; Ast G The Alternative Role of DNA Methylation in Splicing Regulation. Trends Genet. 2015, 31, 274–280. [DOI] [PubMed] [Google Scholar]
- 10.Vakoc CR; Mandat SA; Olenchock BA; et al. Histone H3 Lysine 9 Methylation and HP1γ Are Associated with Transcription Elongation through Mammalian Chromatin. Mol. Cell 2005, 19, 381–391. [DOI] [PubMed] [Google Scholar]
- 11.Canzio D; Chang EY; Shankar S; et al. Chromodomain-Mediated Oligomerization of HP1 Suggests a Nucleosome-Bridging Mechanism for Heterochromatin Assembly. Mol. Cell 2011, 41, 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon MD; Chu F; Racki LR; et al. The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones. Cell 2007, 128, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallrath LL; Vitalini MW; Elgin SCR Heterochromatin: A Critical Part of the Genome In Fundamentals of Chromatin; Workman JL, Abmayr SM, Eds.; Springer: New York, 2014; pp 529–552. [Google Scholar]
- 14.Fritsch L; Robin P; Mathieu JRR; et al. A Subset of the Histone H3 Lysine 9 Methyltransferases Suv39h1, G9a, GLP, and SETDB1 Participate in a Multimeric Complex. Mol. Cell 2010, 37, 46–56. [DOI] [PubMed] [Google Scholar]
- 15.Sampath SC; Marazzi I; Yap KL; et al. Methylation of a Histone Mimic within the Histone Methyltransferase G9a Regulates Protein Complex Assembly. Mol. Cell 2007, 27, 596–608. [DOI] [PubMed] [Google Scholar]
- 16.Rathert P; Dhayalan A; Murakami M; et al. Protein Lysine Methyltransferase G9a Acts on Non-Histone Targets. Nat. Chem. Biol. 2008, 4, 344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J; Dorsey J; Chuikov S; et al. G9a and Glp Methylate Lysine 373 in the Tumor Suppressor p53. J. Biol. Chem. 2010, 285, 9636–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X; Wen H; Shi X Lysine Methylation: Beyond Histones. Acta Biochim. Biophys. Sin. (Shanghai) 2012, 44, 14–27. [DOI] [PubMed] [Google Scholar]
- 19.Tchasovnikarova IA; Timms RT; Matheson NJ; et al. Epigenetic Silencing by the HUSH Complex Mediates Position-Effect Variegation in Human Cells. Science 2015, 348, 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Koning L; Savignoni A; Boumendil C; et al. Heterochromatin Protein 1alpha: A Hallmark of Cell Proliferation Relevant to Clinical Oncology. EMBO Mol. Med. 2009, 1, 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceol CJ; Houvras Y; Jane-Valbuena J; et al. The Histone Methyltransferase SETDB1 Is Recurrently Amplified in Melanoma and Accelerates Its Onset. Nature 2011, 471, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiba T; Saito T; Yuki K; et al. Histone Lysine Methyltransferase SUV39H1 Is a Potent Target for Epigenetic Therapy of Hepatocellular Carcinoma. Int. J. Cancer 2015, 136, 289–298. [DOI] [PubMed] [Google Scholar]
- 23.Hathaway NA; Bell O; Hodges C; et al. Dynamics and Memory of Heterochromatin in Living Cells. Cell 2012, 149, 1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanton BZ; Chory EJ; Crabtree GR Chemically Induced Proximity in Biology and Medicine. Science 2018, 359, eaao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiarella AM; Wang TA; Butler KV; et al. Repressing Gene Transcription by Redirecting Cellular Machinery with Chemical Epigenetic Modifiers. J. Vis. Exp. 2018, 1, e58222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lienert F; Wirbelauer C; Som I; et al. Identification of Genetic Elements That Autonomously Determine DNA Methylation States. Nat. Genet. 2011, 43, 1091–1097. [DOI] [PubMed] [Google Scholar]
- 27.Elling U; Wimmer RA; Leibbrandt A; et al. A Reversible Haploid Mouse Embryonic Stem Cell Biobank Resource for Functional Genomics. Nature 2017, 550, 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ; Schmittgen TD Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-AACT Method. Methods 2001, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 29.Allfrey VG; Faulkner R; Mirsky AE Acetylation and Methylation of Histones and Their Possible Role in the Regulation of RNA Synthesis. Proc. Natl. Acad. Sci. U.S.A. 1964, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y; Lee H-M; Xiong Y; et al. Targeting the Histone Methyltransferase G9a Activates Imprinted Genes and Improves Survival of a Mouse Model of Prader-Willi Syndrome. Nat. Med. 2016, 23, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldman N; Gerson A; Fang J; et al. G9a-Mediated Irreversible Epigenetic Inactivation of Oct-3/4 during Early Embryogenesis. Nat. Cell Biol. 2006, 8, 188–194. [DOI] [PubMed] [Google Scholar]
- 32.Ross PL; Huang YN; Marchese JN; et al. Multiplexed Protein Quantitation in Saccharomyces cerevisiae Using Amine-Reactive Isobaric Tagging Reagents. Mol. Cell. Proteomics 2004, 3, 1154–1169. [DOI] [PubMed] [Google Scholar]
- 33.Wong C-M; Wei L; Law C-T; et al. Up-Regulation of Histone Methyltransferase SETDB1 by Multiple Mechanisms in Hepatocellular Carcinoma Promotes Cancer Metastasis. Hepatology 2016, 63, 474–487. [DOI] [PubMed] [Google Scholar]
- 34.Baude A; Aaes TL; Zhai B; et al. Hepatoma-Derived Growth Factor-Related Protein 2 Promotes DNA Repair by Homologous Recombination. Nucleic Acids Res. 2016, 44, 2214–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H; Jurado KA; Wu X; et al. HRP2 Determines the Efficiency and Specificity of HIV-1 Integration in LEDGF/P75 Knockout Cells but Does Not Contribute to the Antiviral Activity of a Potent LEDGF/P75-Binding Site Integrase Inhibitor. Nucleic Acids Res. 2012, 40, 11518–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.