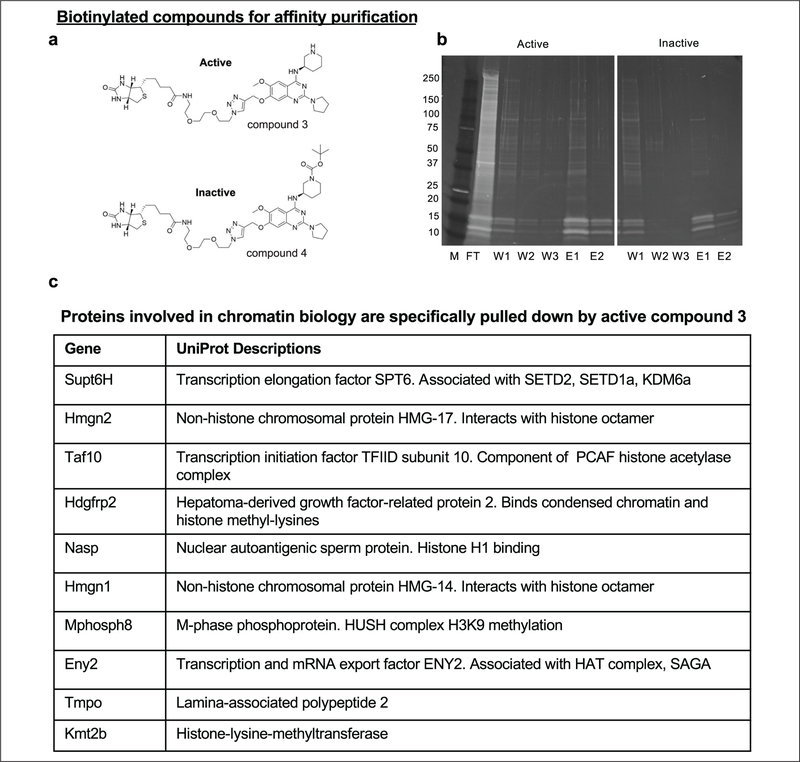
Figure 5.
Chemical proteomics identifies putative HPI-heterochromatin pathway components. (a) Compound 3 (active) and compound 4 (inactive control) were used for chemical affinity purification to identify binding targets of compound 2 from CiA:Oct4 sonicated nuclear lysates. (b) Tris-glycine gradient protein gels (4%–20%) stained with Sypro Ruby stain loaded with marker (M), flow-through (FT), wash 1 (W1), wash 2 (W2), wash 3 (W3), elution 1 (E1), and elution 2 (E2) from affinity pulldown experiments comparing compounds 3 and 4. (c) The table lists select proteins enriched in the compound 3 sample (active) compared with controls as identified by iTRAQ LC-MS/MS with a summary of their UniProt descriptions.