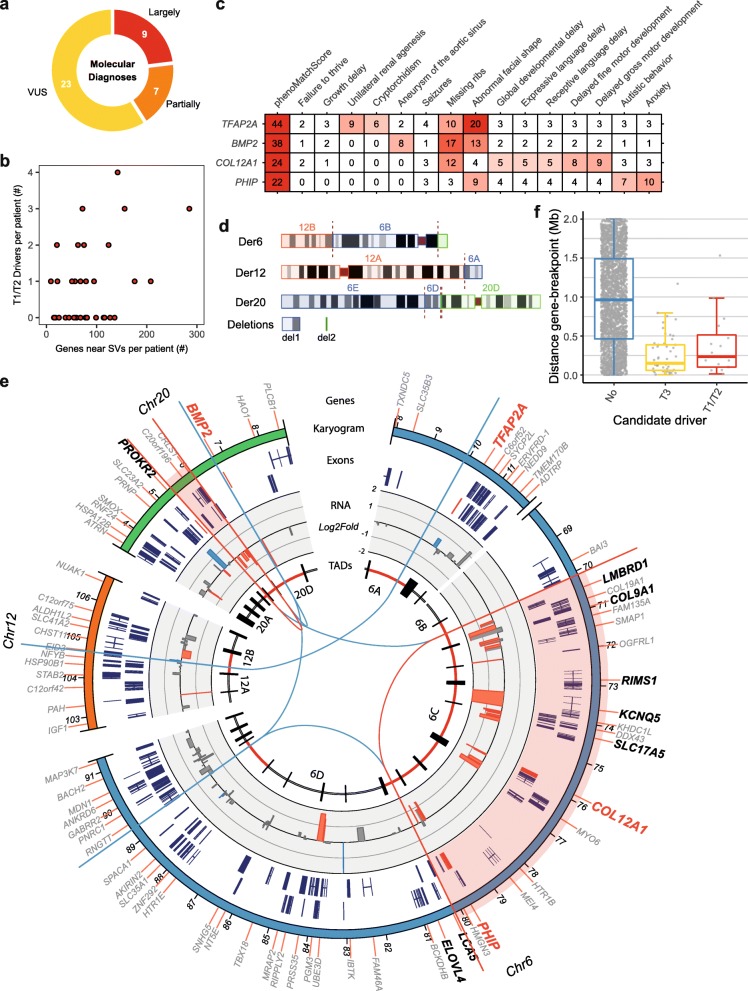
Fig. 3.
SVs can affect multiple candidate drivers which jointly contribute to a phenotype. a Number of patients whose phenotype can be partially or largely explained by the predicted T1/T2 candidate drivers (based on the percentage of the patient’s HPO terms that have a phenomatch score > 4). These molecular diagnoses are based on the fraction of HPO terms assigned to the patients that have a phenomatch score of more than 5 with at least one T1/T2 driver gene. b Scatterplot showing the number of predicted T1/T2 candidate drivers compared to the total number of genes at or adjacent (< 2 Mb) to the de novo SVs per patient. c Heatmap showing the association of the four predicted T1/T2 candidate drivers with the phenotypic features (described by HPO terms) of individual P25. The numbers correspond to the score determined by phenomatch. The four genes are associated with different parts of the complex phenotype of the patient. d Ideogram of the derivative (der) chromosomes 6, 12, and 20 in individual P25 reconstructed from the WGS data. WGS detected complex rearrangements with six breakpoint junctions and two deletions on chr6 and chr20 respectively of ~ 10 Mb and ~ 0.6 Mb. e Circos plot showing the genomic regions and candidate drivers affected by the complex rearrangements in individual P25. Gene symbols of T1/T2 and T3 candidate drivers are shown respectively in red and black. The breakpoint junctions are visualized by the lines in the inner region of the plot (red lines and highlights indicate the deletions). The middle ring shows the log2 fold change RNA expression changes in lymphoblastoid cells derived from the patient compared to controls measured by RNA sequencing. Genes differentially expressed (p < 0.05) are indicated by red (log2 fold change < − 0.5) and blue (log2 fold change > 0.5) bars. The inner ring shows the organization of the TADs and their boundaries (indicated by vertical black lines) in germinal zone (GZ) brain cells [35]. TADs overlapping with the de novo SVs are highlighted in red. f Genomic distance (in base pairs) between the indirectly affected candidate driver genes and the closest breakpoint junction. Most candidate drivers are located within 1 Mb of a breakpoint junction (median distance of 185 kb)