Fig. 4.
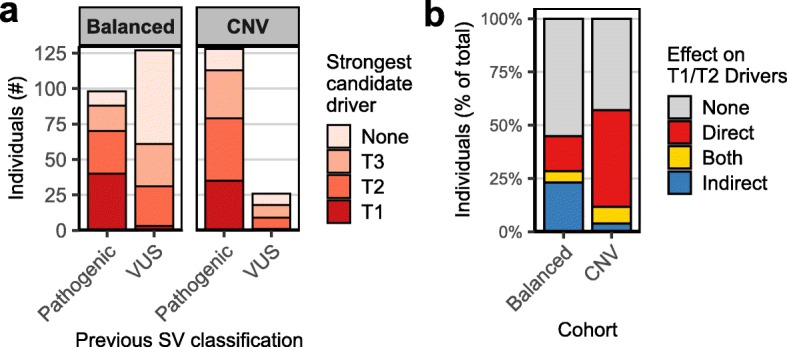
In silico prediction of candidate drivers in larger cohorts of patients with de novo SVs. a Comparison between previous SV classifications with the strongest candidate driver (located at or adjacent (< 1 Mb) to these SVs) predicted by our approach. Two different patient cohorts, one containing mostly balanced SVs [21] and one containing copy number variants, were screened for candidate drivers. Our method identified T1/T2 candidate drivers for most SVs previously classified as pathogenic or likely pathogenic. Additionally, the method detected T1/T2 candidate drivers for some SVs previously classified as VUS, which may lead to a new molecular diagnosis. b Quantification of the predicted effects of the SVs on proposed T1/T2 candidate driver genes per cohort. Individuals with multiple directly and indirectly affected candidate drivers are grouped in the category described as “Both.” Indirect position effects of SVs on genes contributing to phenotypes appear to be more common in patients with balanced SVs compared to patients with copy number variants
