Fig. 3.
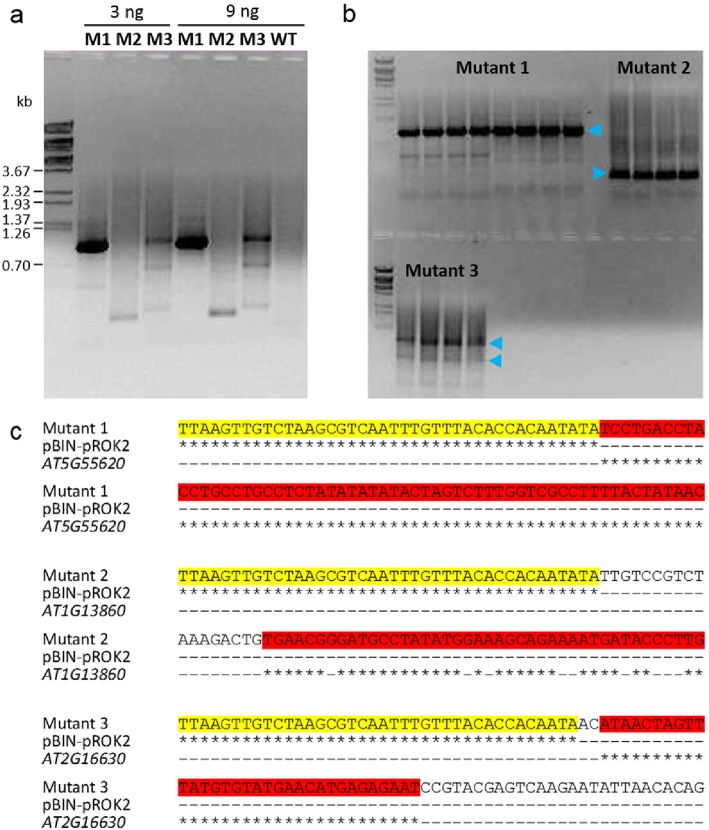
Identification of candidate genes by adapter ligation-mediated PCR. a DNA of the mutants 1, 2, and 3 (M1, M2, M3) was extracted and T-DNA containing fragments were selectively amplified by nested PCR. DNA of a wild-type plant (WT, Col-0) served as T-DNA negative control. For each mutant line 3 and 9 ng DNA was loaded on an agarose gel. DNA standards are indicated. b Multiple PCR reactions (9 ng DNA per lane) were loaded on an agarose gel to obtain sufficient amounts of DNA for sequencing. Major bands (blue arrow heads) were cut, and the eluted DNA was sequenced. c Results of sequencing, homology search and candidate gene alignment are shown. For mutant 3 both cut bands contained DNA with the displayed consensus sequence. Homology of mutant DNA to the pBIN-pROK2 vector (T-DNA) or an Arabidopsis gene is highlighted in yellow and red, respectively. Asterisks indicate matching bases. Only 100 bases around the T-DNA-gene junctions are shown. BLAST hit for Mutant 1: NM_124944.4, AT5G55620, E value 0.0, 100% identities (726/726). BLAST hit for Mutant 2: CP002684.1, AT1G13860, QUL1, E value 0.0, 89% identities (527/592). BLAST hit for Mutant 3: NM_127215.5, AT2G16630, FOCL1, E value 2e-06, 100% identities (33 of 33). The T-DNA-flanking genomic sequences used for the BLAST searches are given in Additional file 2: Table S1