Fig. 1.
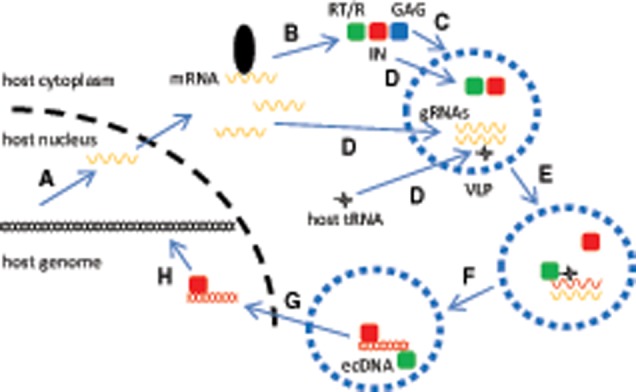
—Simplified classical representation of LTR retrotransposons life-cycle. (A) LTR retrotransposon is activated transcriptionally (black strand, host DNA; yellow strand, TE messenger/genomic RNA [mRNA/gRNA]). (B) mRNA is translated and cleaved into functional proteins, including a reverse-transcriptase/RNaseH (RT/R, green box), an integrase (IN, red box) and a structural GAG (blue box). (C) GAG assembles as virus-like particles (VLP) within the host cytoplasm. (D) Two gRNA molecules are copackaged along with proteins and host tRNA. (E) tRNA anneals the primer-binding-site, and primed RT/R eventually synthetizes a linear double-stranded extrachromosomal DNA (ds-ecDNA, red strand) intermediary, using gRNAs as templates. (F and G) ds-ecDNA associates with the integrase and migrates into the nucleus using yet unidentified mechanisms. (H) ds-ecDNA intermediary eventually inserts into a new location within the host genome, resulting in a new LTR retrotransposon copy.
