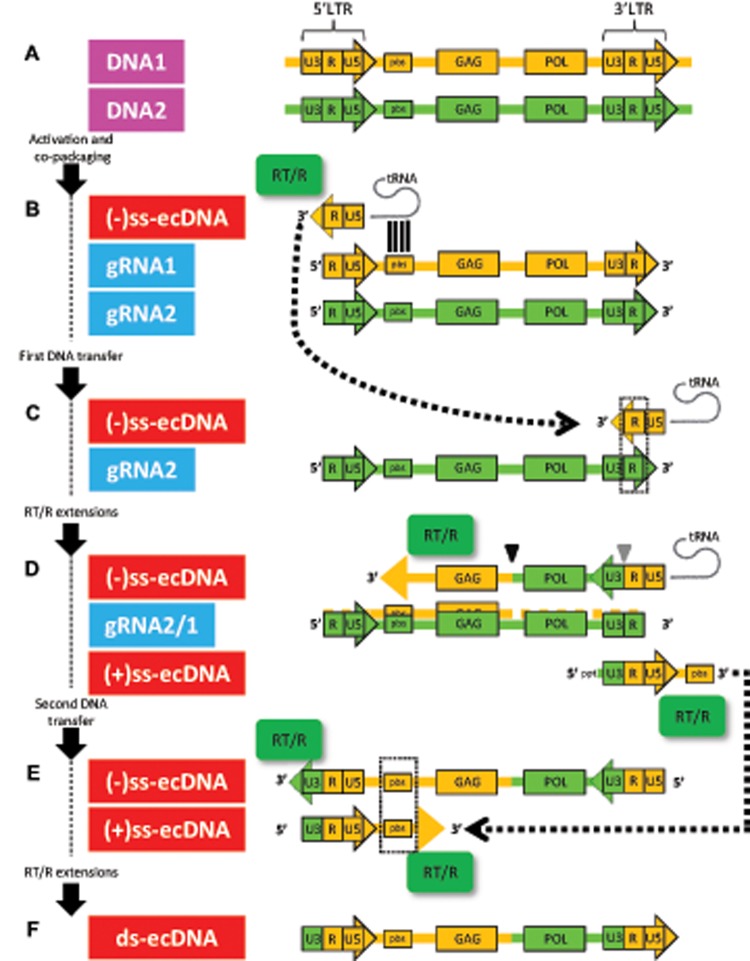
Fig. 2.
—Schematic representation of discontinuous reverse-transcription and recombination steps. (A) Two members of a LTR retrotransposon clan are transcriptionally activated and the corresponding genomic RNA (gRNA) progenitors (starting and ending in the R regions of 5′- and 3′-LTR, respectively) are copackaged. (B) Reverse-transcriptase/RNaseH activity (RT/R, green) is primed by tRNA annealing the priming-binding-site (pbs) of gRNA1 (orange), and minus-single-strand extrachromosomal cDNA ((-)ss-ecDNA) is synthesized. (C) First strand transfer: strong-stop (-)ss-ecDNA transfers to a second gRNA2 (green) (first recombinogenic step; the hypothesized recombination point is marked with an inverted gray triangle), using sequence homologies in the R region (marked with a dotted box). (D) At the same time that RNAseH activity proceeds (not shown), the (-)ss-ecDNA is extended using gRNAs as alternate templates (second recombinogenic step; a single hypothesized recombination point is marked with an inverted black triangle, but note that more than one event is possible). The color-coded newly synthesized (-)ss-ecDNA molecule is represented as a mosaic of the two progenitor gRNAs. Although RNAseH activity degrades portions of gRNA2 template (not shown), priming of poly-purine-track (ppt) allows nascent plus-single-strand extrachromosomal cDNA ((+)ss-ecDNA) synthesis until the end of the (-)ss-ecDNA molecule used as template. (E) Second strand transfer: strong-stop (+)ss-ecDNA swaps toward the 5′ area of the (-)ss-ecDNA. In addition, RT/R final extensions take place. (F) After final extensions, a mosaic blunt-ended linear extrachromosomal DNA (ds-ecDNA) molecule with two identical LTRs is generated. U3/R/U5, domains within LTRs; GAG, structural protein; POL, polyprotein.