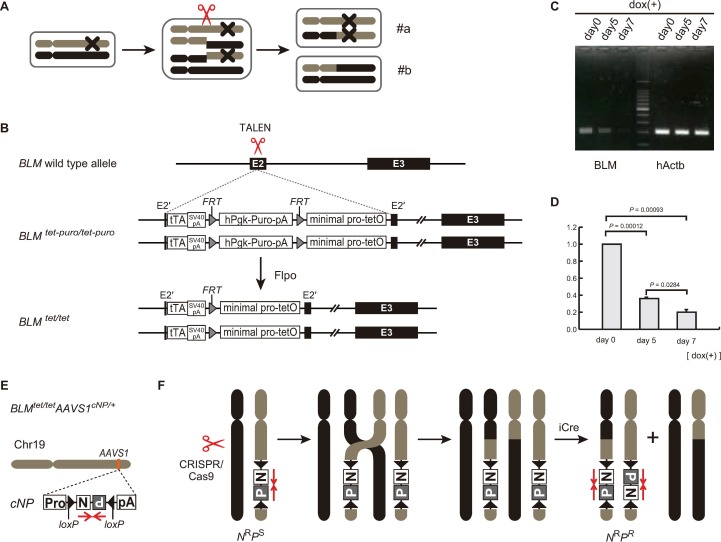
Fig 1. Establishment of a BLM transcript regulation system and detection of crossovers by a reporter cassette in hiPSCs.
(A) Schematic representation of a crossover induced by a double-stranded DNA break during the 4N stage of the cell cycle in hiPSCs. X is a dominant mutation. After segregation, case #a cells have the disease phenotype, while case #b cells are normal. (B) Targeting of the Tet-Off cassette to both alleles of the BLM locus with the assistance of TALEN (BLMtet-puro/tet-puro). The selection cassettes were removed by Flippase (Flpo), the hiPSC-BLMtet/tet cells were established. (C) Electrophoresis of qRT-PCR products. Total RNA from 2 × 105 hiPSC-BLMtet/tet cells was collected at days 0, 5, and 7 after dox treatment, and qRT-PCR was performed. (D) Histogram data represent the mean ± SEM of three independent experiments. Comparison of two groups with normally distributed variables was performed using the Student’s t-test by Kaleida Graph software. (E) Targeting of the double selection cassette (cNP) to the AAVS1 locus (hiPSC-BLMtet/tetAAVScNP/+). (F) Schematic representation of mitotic crossover during the 4N stage. DSBs were introduced by CRISPR/Cas9.