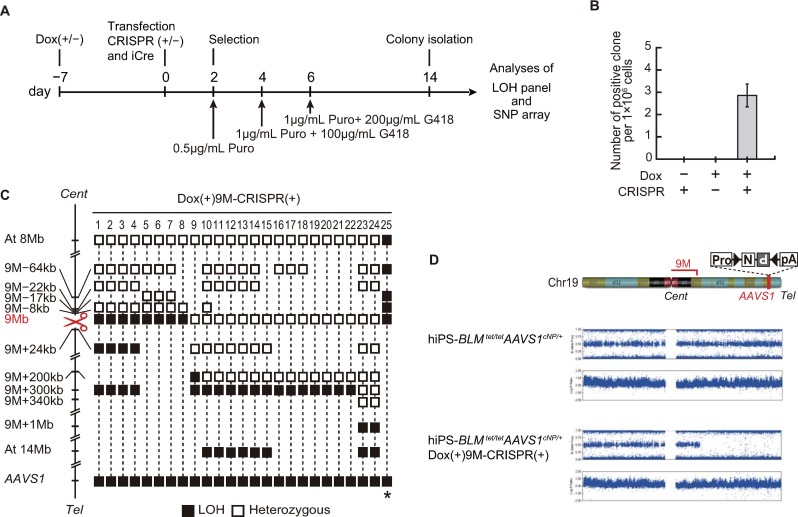
Fig 2. Generation and analyses of clones with crossovers in chromosome 19.
Procedure for the isolation of NRPR clones under the condition of BLM transcript suppression. hiPSC-BLMtet/tetAAVS1cNP/+ cells were treated with or without dox from 7 days before transfection to the day of transfection. Selection started 2 days after transfection, either with or without the introduction of an allele-specific DSB. (B) Efficiency of crossover under the various conditions at the 9 Mb region of chromosome 19. We picked 439 colonies (eight trials) and performed genotyping (S5 Fig). Bar indicates SEM. (C) Distribution of crossover points at 9 Mb from the centromere of chromosome 19. Twenty-five NRPR-positive clones were isolated and analyzed using LOH panels and SNP arrays. Open squares indicate the heterozygous genotype. Closed black squares indicate the loss of heterozygosity (LOH) genotype. The DSBs introduced by the CRISPR/Cas9 system are indicated by scissors. (D) A schematic representation of chromosome 19 is shown at the top of the figure. After crossover, SNP array analysis indicated that the genome derived from hiPSC-BLMtet/tetAAVS1cNP/+ Dox(+)9M-CRISPR(+) cells showed homozygosity telomeric to DSB and that the cells maintained two copies of the genome.